| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,208,232 – 7,208,340 |
| Length | 108 |
| Max. P | 0.965511 |

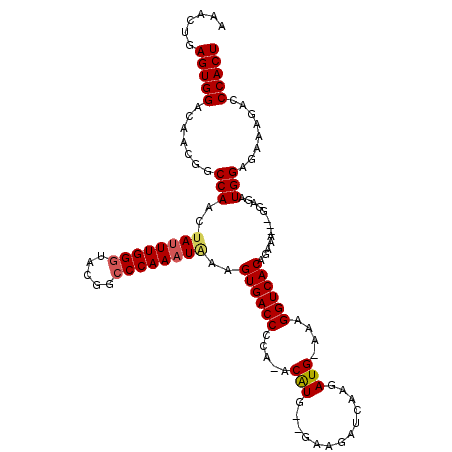
| Location | 7,208,232 – 7,208,340 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -29.41 |
| Consensus MFE | -25.02 |
| Energy contribution | -24.47 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7208232 108 + 23771897 AGUGGGUCUUUCACCAUA------UUUCUGUGACCUUU-CAUCUCGAUCUGCCCCACGUUUUGGGUCACUCUAGUUGGGCCGAAUCCAAUUAGGUGGCUGUUGUCCACUCAGUUU ((((((.(...((((((.------.....(((((((..-.....((..........))....))))))).(((((((((.....))))))))))))).))..)))))))...... ( -30.10) >DroEre_CAF1 21671 111 + 1 AGUGGGUCUUUCUCCAUCUCCACUUUUCUGUGACCUUU-CAUCUUGAUCUUC--CAUGU-UGGGGUCACUUCAUUUGGGCCGUACCCAAAUAGUUGGCCGUUGUCCACUCAGUUU ((((((.(.....(((.((..........((((((((.-...(.((......--)).).-.))))))))...(((((((.....))))))))).))).....)))))))...... ( -27.80) >DroYak_CAF1 18952 112 + 1 AGUGGCUCUUUCUCCAUCUCCCUGCUACUGUGACCUUUCCAUCUUGAUCUUC--CAUGU-UGGGGUCACUUUAUUUGGGCCAUACCCAAAUGGUUGGCCAUUGUCCACUCAGUUU ((((((.................))))))((((((((..(((..........--.))).-.))))))))........(((((.(((.....))))))))((((......)))).. ( -30.33) >consensus AGUGGGUCUUUCUCCAUCUCC___UUUCUGUGACCUUU_CAUCUUGAUCUUC__CAUGU_UGGGGUCACUUUAUUUGGGCCGUACCCAAAUAGUUGGCCGUUGUCCACUCAGUUU ((((((.(..(..(((.............(((((((..........................))))))).(((((((((.....))))))))).)))..)..)))))))...... (-25.02 = -24.47 + -0.55)

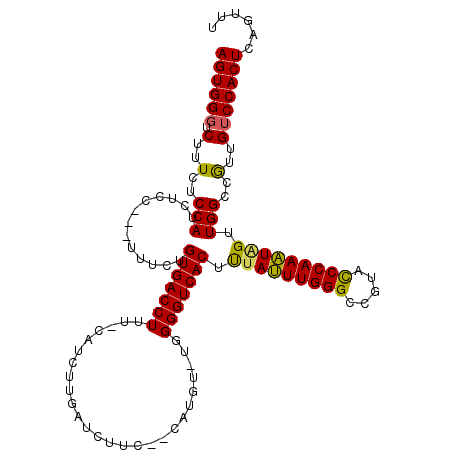
| Location | 7,208,232 – 7,208,340 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -23.01 |
| Energy contribution | -23.68 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7208232 108 - 23771897 AAACUGAGUGGACAACAGCCACCUAAUUGGAUUCGGCCCAACUAGAGUGACCCAAAACGUGGGGCAGAUCGAGAUG-AAAGGUCACAGAAA------UAUGGUGAAAGACCCACU ......(((((......((((((.....))....((((((.((.((.((.((((.....)))).))..)).)).))-...)))).......------..)))).......))))) ( -24.92) >DroEre_CAF1 21671 111 - 1 AAACUGAGUGGACAACGGCCAACUAUUUGGGUACGGCCCAAAUGAAGUGACCCCA-ACAUG--GAAGAUCAAGAUG-AAAGGUCACAGAAAAGUGGAGAUGGAGAAAGACCCACU ......(((((.......(((.(((((((((.....)))))))...((((((((.-....)--)....((.....)-)..))))))..........)).)))........))))) ( -28.86) >DroYak_CAF1 18952 112 - 1 AAACUGAGUGGACAAUGGCCAACCAUUUGGGUAUGGCCCAAAUAAAGUGACCCCA-ACAUG--GAAGAUCAAGAUGGAAAGGUCACAGUAGCAGGGAGAUGGAGAAAGAGCCACU ......(((((.......(((.(((((((((.....)))))))...(((((((((-.(.((--......)).).)))...)))))).......))....)))........))))) ( -32.16) >consensus AAACUGAGUGGACAACGGCCAACUAUUUGGGUACGGCCCAAAUAAAGUGACCCCA_ACAUG__GAAGAUCAAGAUG_AAAGGUCACAGAAA___GGAGAUGGAGAAAGACCCACU ......(((((.......(((..((((((((.....))))))))..((((((.....(((.............)))....)))))).............)))........))))) (-23.01 = -23.68 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:05 2006