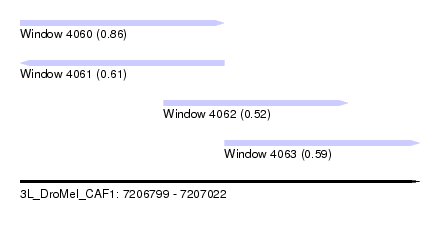
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,206,799 – 7,207,022 |
| Length | 223 |
| Max. P | 0.864540 |

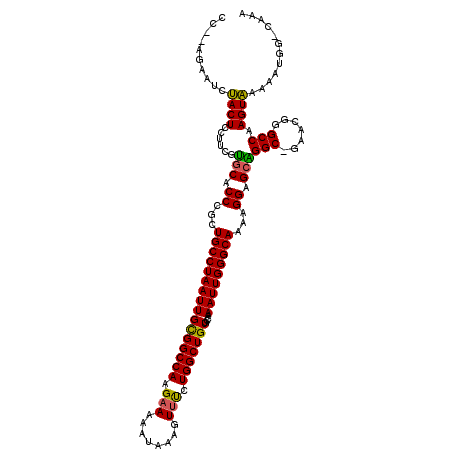
| Location | 7,206,799 – 7,206,913 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.43 |
| Mean single sequence MFE | -38.93 |
| Consensus MFE | -28.20 |
| Energy contribution | -27.43 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7206799 114 + 23771897 CCCGAGAAUUCACUCCUUUGGCCACCCGCUGCCUAAUUGCGGCCAAGAAAAAUAAAGUUUCUGGCUGUGCCAAAUUGGGCAAAAGGAGCUGGC-GAACGGGCCAAGUGAAAAUGG----- ........(((((((((((.(((((((..((((((((((((((((.((((.......)))))))))))....)))))))))...)).).))))-.)).)))...)))))).....----- ( -40.70) >DroEre_CAF1 20345 118 + 1 CC--AGAAUCUACUACUCCGUGCACCCGCUGCCUAAUUGUGGCCAAGAAAAAUAAAGUUCCUGGCUGUGCCAAAUUGGGCAAAAGGAGCAGGCAGAACGGGCCAAGUAAGAAUGGGCAAA ((--(...((((((.((((..((....))((((((((((..((((.(((........))).))))..)....)))))))))...))))..(((.......))).))).))).)))..... ( -35.90) >DroYak_CAF1 17659 117 + 1 CC--AGAAACUACUCCUUCGUGCACCCGCUGCCUAAUUGCGGCCAAGAAAAAUAAGUUCUCUGGCUGUGCCAAAUUGGGCAAAAGGAGCGGGC-GAAUGGGCCAAGUAAAAAUGGACAAA ((--(.....((((..(((((.(.((((((.(((....(((((((((((.......)))).)))))))(((......)))...))))))))).-).)))))...))))....)))..... ( -40.20) >consensus CC__AGAAUCUACUCCUUCGUGCACCCGCUGCCUAAUUGCGGCCAAGAAAAAUAAAGUUUCUGGCUGUGCCAAAUUGGGCAAAAGGAGCAGGC_GAACGGGCCAAGUAAAAAUGG_CAAA ..........((((......(((.((...((((((((((((((((.(((........))).)))))))....)))))))))...)).)))(((.......))).))))............ (-28.20 = -27.43 + -0.77)

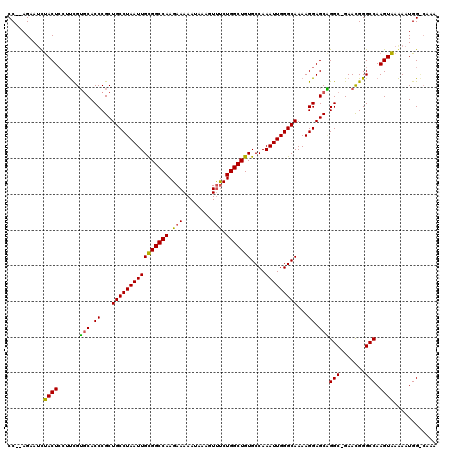
| Location | 7,206,799 – 7,206,913 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.43 |
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -29.66 |
| Energy contribution | -29.00 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7206799 114 - 23771897 -----CCAUUUUCACUUGGCCCGUUC-GCCAGCUCCUUUUGCCCAAUUUGGCACAGCCAGAAACUUUAUUUUUCUUGGCCGCAAUUAGGCAGCGGGUGGCCAAAGGAGUGAAUUCUCGGG -----(((........)))((((...-....((((((((.((((..((((((...)))))).................((((.........))))).))).)))))))).......)))) ( -36.44) >DroEre_CAF1 20345 118 - 1 UUUGCCCAUUCUUACUUGGCCCGUUCUGCCUGCUCCUUUUGCCCAAUUUGGCACAGCCAGGAACUUUAUUUUUCUUGGCCACAAUUAGGCAGCGGGUGCACGGAGUAGUAGAUUCU--GG .....(((.(((.(((..((((((.(.((((((((((..((((......))))..((((((((........)))))))).......))).)))))))).)))).))))))))...)--)) ( -40.00) >DroYak_CAF1 17659 117 - 1 UUUGUCCAUUUUUACUUGGCCCAUUC-GCCCGCUCCUUUUGCCCAAUUUGGCACAGCCAGAGAACUUAUUUUUCUUGGCCGCAAUUAGGCAGCGGGUGCACGAAGGAGUAGUUUCU--GG .....(((...((((((........(-((((((((((.((((((.....))....(((((((((.....))))).)))).))))..))).)))))))).......))))))....)--)) ( -36.16) >consensus UUUG_CCAUUUUUACUUGGCCCGUUC_GCCAGCUCCUUUUGCCCAAUUUGGCACAGCCAGAAACUUUAUUUUUCUUGGCCGCAAUUAGGCAGCGGGUGCACGAAGGAGUAGAUUCU__GG ...........(((((((((.......))).((.(((.(((((.((((((((.((...(((((.......))))))))))).)))).))))).))).))......))))))......... (-29.66 = -29.00 + -0.66)

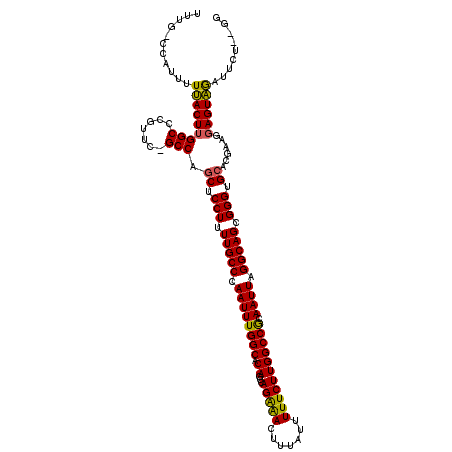
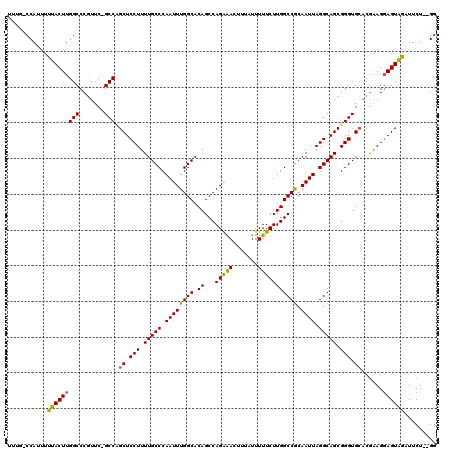
| Location | 7,206,879 – 7,206,982 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.29 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -18.72 |
| Energy contribution | -19.17 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7206879 103 + 23771897 AAAAGGAGCUGGC-GAACGGGCCAAGUGAAAAUGG-------------GGAAAAACAUGUGCAAUGUCAUAGGACUGAGCAUGAGAUACGGCAAUGUCGGCAU---GAAACAAGCUUUUG ...((((((((((-......)))..((....(((.-------------.......(((((.....(((....)))...))))).....((((...)))).)))---...)).))))))). ( -23.40) >DroEre_CAF1 20423 120 + 1 AAAAGGAGCAGGCAGAACGGGCCAAGUAAGAAUGGGCAAAGACGGCUGGGAAAAACAUGUGCAAUGUCGUAUGACAGAGCAUGAGAUACGGCAAUGUUGCAAUGUUGAAACAAGCUUUUG ...((((((.(((......(.(((........))).).......))).(....(((((.(((((((((((((..((.....))..))))))))...))))))))))....)..)))))). ( -30.62) >DroYak_CAF1 17737 119 + 1 AAAAGGAGCGGGC-GAAUGGGCCAAGUAAAAAUGGACAAAGACGGCUGGGAAAAACGUGUGCAAUGUCGUAUGGCAGAGCAUGAGAUACGGCAAUGUUGCAAUGUUGAAACAAGCUUUUG ...((((((((.(-(..(..((((........))).)..)..)).)).(....(((((.(((((((((((((..((.....))..))))))))...))))))))))....)..)))))). ( -30.40) >consensus AAAAGGAGCAGGC_GAACGGGCCAAGUAAAAAUGG_CAAAGACGGCUGGGAAAAACAUGUGCAAUGUCGUAUGACAGAGCAUGAGAUACGGCAAUGUUGCAAUGUUGAAACAAGCUUUUG ...((((((.(((.......)))..............................(((((.(((..((((.((((......)))).))))..))))))))...............)))))). (-18.72 = -19.17 + 0.45)

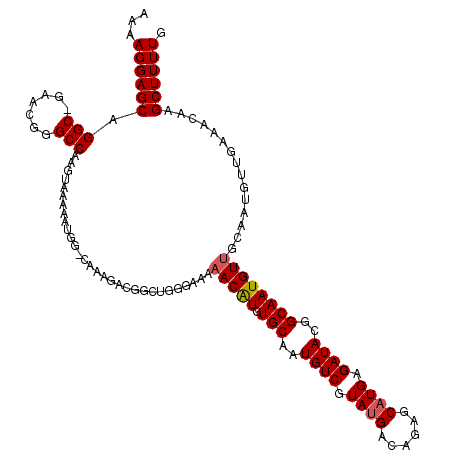
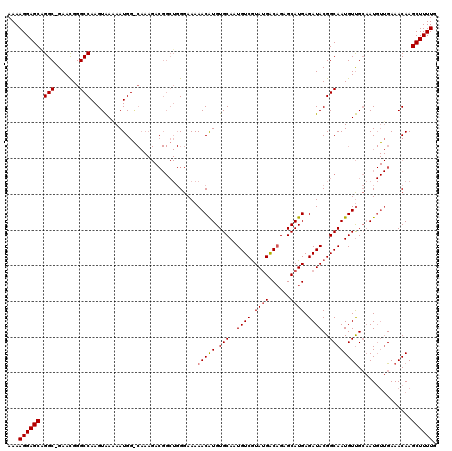
| Location | 7,206,913 – 7,207,022 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -29.81 |
| Consensus MFE | -21.37 |
| Energy contribution | -23.93 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7206913 109 + 23771897 --------GGAAAAACAUGUGCAAUGUCAUAGGACUGAGCAUGAGAUACGGCAAUGUCGGCAU---GAAACAAGCUUUUGCCAAAAAUAUUGCAAACAAGUGCACAUAUGCAACCAUAAA --------((......(((((((..(((....)))...(((........(((((....(((.(---(...)).))).)))))........))).......)))))))......))..... ( -22.99) >DroEre_CAF1 20463 120 + 1 GACGGCUGGGAAAAACAUGUGCAAUGUCGUAUGACAGAGCAUGAGAUACGGCAAUGUUGCAAUGUUGAAACAAGCUUUUGCCAAAAAUAUUGCAAGCAACUGCACAUAUGCAACCAUAAA ......(((.......(((((((.((((((((..((.....))..)))))))).((((((((((((.......((....))....)))))))).))))..)))))))......))).... ( -32.32) >DroYak_CAF1 17776 120 + 1 GACGGCUGGGAAAAACGUGUGCAAUGUCGUAUGGCAGAGCAUGAGAUACGGCAAUGUUGCAAUGUUGAAACAAGCUUUUGCCAAAAAUAUUGCAAGCAAGUGCACAUAUGCAACCAUAAA ......(((.......(((((((.((((((((..((.....))..)))))))).((((((((((((.......((....))....)))))))).))))..)))))))......))).... ( -34.12) >consensus GACGGCUGGGAAAAACAUGUGCAAUGUCGUAUGACAGAGCAUGAGAUACGGCAAUGUUGCAAUGUUGAAACAAGCUUUUGCCAAAAAUAUUGCAAGCAAGUGCACAUAUGCAACCAUAAA ........((......(((((((.((((((((..((.....))..)))))))).((((((((((((.......((....))....)))))))).))))..)))))))......))..... (-21.37 = -23.93 + 2.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:03 2006