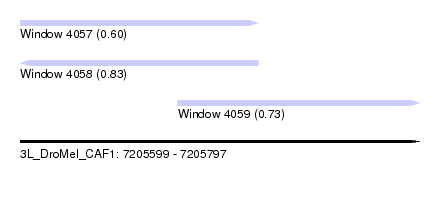
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,205,599 – 7,205,797 |
| Length | 198 |
| Max. P | 0.833617 |

| Location | 7,205,599 – 7,205,717 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.41 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -24.47 |
| Energy contribution | -25.96 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.600791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7205599 118 + 23771897 CUGCCCUUUGAACUUCACAACCGGGCCACAUUUACACAUUUACAUGGCCA--UCGCGAUAACCAUCUCAACGGCGUUUCUCACCUGAGAAAUGGCCCCGGAUCCGGUGGGCUCCUUUGCG ..((((..((.....))..((((((((................((.((..--..)).))............((((((((((....))))))).)))..)).))))))))))......... ( -32.20) >DroSec_CAF1 18892 118 + 1 CUGCCCUUUGAACUUCGCAACCGGGCCACAUUUACACAUUUACAUGGCCA--UCGUGAUACCCAUCUCAACGGCGUUUCUCACCUGAGAAAUGACCCCGGAUCCGGCGGUCUCCUUUGCG ...............(((((((((((((................))))).--..(.(((....))).)..))).(((((((....)))))))(((((((....))).))))....))))) ( -33.69) >DroEre_CAF1 19315 110 + 1 CUGCCCUUUGAACUUCGCACCCGGCCCACAU--------UUACAUGGGCA--ACGCGAUUCCCAUCUCCUCGGCGUUCCUCACCUGAGAAAUGGCCCCGGAUACCGGGGUCUCCUUUUCG ..(((....((...((((.....(((((...--------.....))))).--..))))......)).....)))...........(((((..((((((((...))))))))...))))). ( -35.10) >DroYak_CAF1 16531 111 + 1 CUGCCCUUUGAACUUCGCACCCGGCCGACAU--------UUACAUGGGCACCACGCGAUACCCAUCUCCACGGCGUUCCUCACCUGAGAAAUGGCCCCGGAU-CCGGGGUCUCCUUUGCG .(((((..((((..((((.....).)))..)--------)))...)))))...(((((......((((...((.(.....).)).))))...(((((((...-.)))))))....))))) ( -35.00) >consensus CUGCCCUUUGAACUUCGCAACCGGCCCACAU________UUACAUGGCCA__ACGCGAUACCCAUCUCAACGGCGUUCCUCACCUGAGAAAUGGCCCCGGAUCCCGGGGUCUCCUUUGCG ..(((....((...((((.....(((((................))))).....))))......)).....)))(((((((....)))))))((((((((...))))))))......... (-24.47 = -25.96 + 1.50)

| Location | 7,205,599 – 7,205,717 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.41 |
| Mean single sequence MFE | -43.92 |
| Consensus MFE | -32.79 |
| Energy contribution | -32.60 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7205599 118 - 23771897 CGCAAAGGAGCCCACCGGAUCCGGGGCCAUUUCUCAGGUGAGAAACGCCGUUGAGAUGGUUAUCGCGA--UGGCCAUGUAAAUGUGUAAAUGUGGCCCGGUUGUGAAGUUCAAAGGGCAG .........((((.(((....)))(((((((((...((((.....))))...))))))))).((((((--(((((((((..........))))))))..)))))))........)))).. ( -46.80) >DroSec_CAF1 18892 118 - 1 CGCAAAGGAGACCGCCGGAUCCGGGGUCAUUUCUCAGGUGAGAAACGCCGUUGAGAUGGGUAUCACGA--UGGCCAUGUAAAUGUGUAAAUGUGGCCCGGUUGCGAAGUUCAAAGGGCAG (((((....((((.(((....)))))))...(((((((((.....))))..)))))(((((..((((.--..(((((....))).))...))))))))).)))))..(((.....))).. ( -39.60) >DroEre_CAF1 19315 110 - 1 CGAAAAGGAGACCCCGGUAUCCGGGGCCAUUUCUCAGGUGAGGAACGCCGAGGAGAUGGGAAUCGCGU--UGCCCAUGUAA--------AUGUGGGCCGGGUGCGAAGUUCAAAGGGCAG ...........((((((...))))))((((((((..((((.....))))..))))))))...(((((.--.((((((....--------..))))))....))))).(((.....))).. ( -43.60) >DroYak_CAF1 16531 111 - 1 CGCAAAGGAGACCCCGG-AUCCGGGGCCAUUUCUCAGGUGAGGAACGCCGUGGAGAUGGGUAUCGCGUGGUGCCCAUGUAA--------AUGUCGGCCGGGUGCGAAGUUCAAAGGGCAG ((((..(....)(((((-.((((.(((..((((((....)))))).))).)))).(((((((((....)))))))))....--------.......)))))))))..(((.....))).. ( -45.70) >consensus CGCAAAGGAGACCCCCGGAUCCGGGGCCAUUUCUCAGGUGAGAAACGCCGUGGAGAUGGGUAUCGCGA__UGCCCAUGUAA________AUGUGGCCCGGGUGCGAAGUUCAAAGGGCAG ......(((.((....)).)))...(((...((((.((((.....))))...))))((((..((((((...((((((((..........))))))))...))))))..))))...))).. (-32.79 = -32.60 + -0.19)

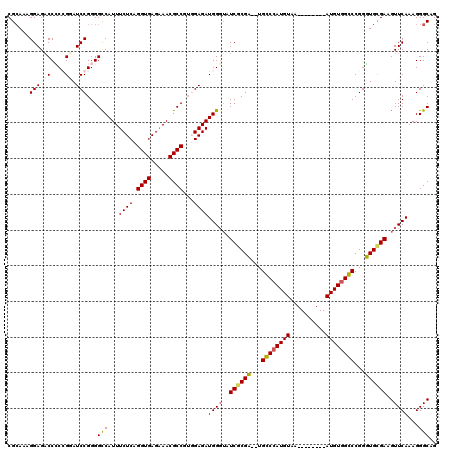
| Location | 7,205,677 – 7,205,797 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.46 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -31.30 |
| Energy contribution | -32.93 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7205677 120 + 23771897 CACCUGAGAAAUGGCCCCGGAUCCGGUGGGCUCCUUUGCGCUCCGGUUACGUGGGGCUGUGUGGGUCCACAAUUUAUCAUGACACUAAUGAGCGUGAAAUAGGACAAGGUUUAUUAAAUU .((((.......(.(((((....))).)).)(((((..(((((.(((..(((((.....((((....)))).....)))))..)))...)))))..)...))))..)))).......... ( -37.00) >DroSec_CAF1 18970 120 + 1 CACCUGAGAAAUGACCCCGGAUCCGGCGGUCUCCUUUGCGCUCCGGUUACGUGGGGCUGUGUGGCUCCACAAUUUAUCAUGACACUAAUGAGCGUGAAAUAGGACAAGGUUUAUUAAAUU .((((.......(((((((....))).))))(((((..(((((.(....)((((((((....))))))))...................)))))..)...))))..)))).......... ( -41.70) >DroEre_CAF1 19385 116 + 1 CACCUGAGAAAUGGCCCCGGAUACCGGGGUCUCCUUUUCGCUCCGGUUACGUGA----CUGUGGCUCCACAAUUUAUCAUGACACUAAUGAGCAUGAAAUAGGACAAGGUUUAUUAAAUU .((((.......((((((((...))))))))((((((((((((.(((..(((((----.((((....)))).....)))))..)))...))))..)))).))))..)))).......... ( -36.60) >DroYak_CAF1 16603 115 + 1 CACCUGAGAAAUGGCCCCGGAU-CCGGGGUCUCCUUUGCGCUCCGGUUACGUGA----CUGUGGCUCCACAAUUUAUCAUGACACUAAUGAGCAUGAAAUAGGACAAGGUUUAUUAAAUU .((((.......(((((((...-.)))))))((((.(((((..(((((....))----)))..))...........((((.......)))))))......))))..)))).......... ( -34.30) >consensus CACCUGAGAAAUGGCCCCGGAUCCCGGGGUCUCCUUUGCGCUCCGGUUACGUGA____CUGUGGCUCCACAAUUUAUCAUGACACUAAUGAGCAUGAAAUAGGACAAGGUUUAUUAAAUU .((((.......((((((((...))))))))((((((((((((.(((..(((((.....((((....)))).....)))))..)))...))))))))...))))..)))).......... (-31.30 = -32.93 + 1.63)

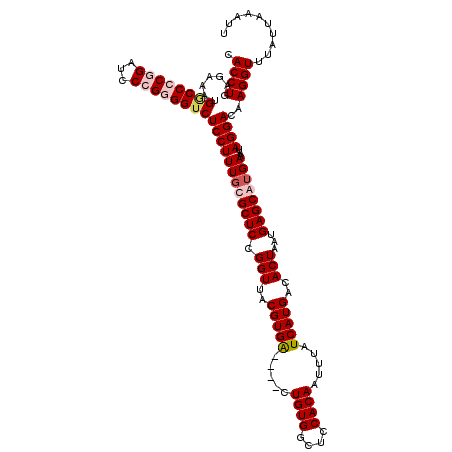
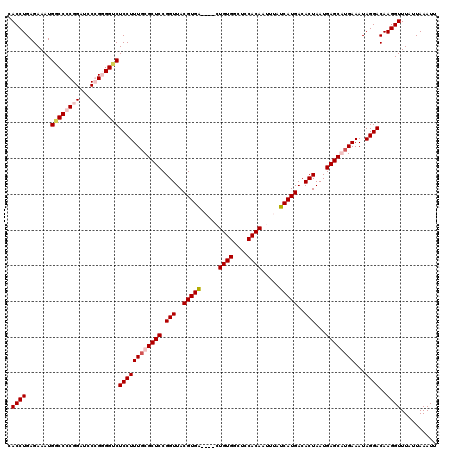
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:00 2006