
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,204,234 – 7,204,657 |
| Length | 423 |
| Max. P | 0.994763 |

| Location | 7,204,234 – 7,204,352 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.13 |
| Mean single sequence MFE | -42.18 |
| Consensus MFE | -30.27 |
| Energy contribution | -31.78 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7204234 118 + 23771897 GGGUGAGGG--GGGUGGACAUUUUAACAACCCAGGAUCGCAGGAUACUAGAAUCCCAGCAUUGCAACAUCCCAGCUUUCCAACAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGC ..((((...--((((.............))))....)))).((((......))))((((((((((((...((((............)))).((((.....)))).)))).)))))))).. ( -38.22) >DroSec_CAF1 17617 120 + 1 GGGUGGGGAGUGGGGGGGCAUUUUAACAACCCAGGAUCGCAGGAUCCUAGAAUCCCAGCAGUGCAACAUCCCAGCUUUCCAGCAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGC ((((..((((((......))))))....))))((((((....))))))......((((((..(((((......(((....)))....(((((((....))))))))))))...)))))). ( -47.10) >DroEre_CAF1 18076 102 + 1 GG----------GGUGGGCAUUUUAACAACCCAGGAUCGCAGCAUCCUGGAAUCCCA--------ACAUCCCAGCUUCCCAGCAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGC ((----------((((((.(((........(((((((......))))))))))))))--------...))))......(((((((.((((((((....))))))))......))))))). ( -39.30) >DroYak_CAF1 15231 110 + 1 GG----------GAUGGGCAUUUUAACAACCCAGGAUCGCUGGAUCCUGGAAUCCCAACAACCCAACAUCCCAGCUUCACAGCAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGC ((----------(.((((.(((........((((((((....)))))))))))))))....)))......(((((......(((..((((((((....)))))))).)))....))))). ( -44.10) >consensus GG__________GGUGGGCAUUUUAACAACCCAGGAUCGCAGGAUCCUAGAAUCCCAGCA_UGCAACAUCCCAGCUUCCCAGCAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGC ...............(((.(((........((((((((....))))))))))))))..............(((((......(((..((((((((....)))))))).)))....))))). (-30.27 = -31.78 + 1.50)



| Location | 7,204,234 – 7,204,352 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.13 |
| Mean single sequence MFE | -41.67 |
| Consensus MFE | -37.12 |
| Energy contribution | -37.00 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7204234 118 - 23771897 GCCAGCAUUGGCAACUGGCAACUUGUGCCGCCAGGAUGUUGGAAAGCUGGGAUGUUGCAAUGCUGGGAUUCUAGUAUCCUGCGAUCCUGGGUUGUUAAAAUGUCCACCC--CCCUCACCC .(((((((((((....((((.....))))))))..))))))).......((((((((((((.(..(((((.(((....))).)))))..))))))...)))))))....--......... ( -38.20) >DroSec_CAF1 17617 120 - 1 GCCAGCAUUGGCAACUGGCAACUUGUGCCGCCAGGAUGCUGGAAAGCUGGGAUGUUGCACUGCUGGGAUUCUAGGAUCCUGCGAUCCUGGGUUGUUAAAAUGCCCCCCCACUCCCCACCC .(((((((((((....((((.....))))))))..))))))).....((((..((......))((((.....((((((....))))))((((.........)))).))))...))))... ( -41.90) >DroEre_CAF1 18076 102 - 1 GCCAGCAUUGGCAACUGGCAACUUGUGCCGCCAGGAUGCUGGGAAGCUGGGAUGU--------UGGGAUUCCAGGAUGCUGCGAUCCUGGGUUGUUAAAAUGCCCACC----------CC .(((((((((((....((((.....))))))))..))))))).....((((...(--------(((.(..(((((((......)))))))..).))))....))))..----------.. ( -40.40) >DroYak_CAF1 15231 110 - 1 GCCAGCAUUGGCAACUGGCAACUUGUGCCGCCAGGAUGCUGUGAAGCUGGGAUGUUGGGUUGUUGGGAUUCCAGGAUCCAGCGAUCCUGGGUUGUUAAAAUGCCCAUC----------CC ....(((((.(((((.((((.....)))).(((((((((((.((..(((((((.(..(....)..).)))))))..)))))).))))))))))))...))))).....----------.. ( -46.20) >consensus GCCAGCAUUGGCAACUGGCAACUUGUGCCGCCAGGAUGCUGGAAAGCUGGGAUGUUGCA_UGCUGGGAUUCCAGGAUCCUGCGAUCCUGGGUUGUUAAAAUGCCCACC__________CC ....(((((.(((((.((((.....)))).(((((((((.(((...(((((((.((........)).)))))))..))).)).))))))))))))...)))))................. (-37.12 = -37.00 + -0.12)

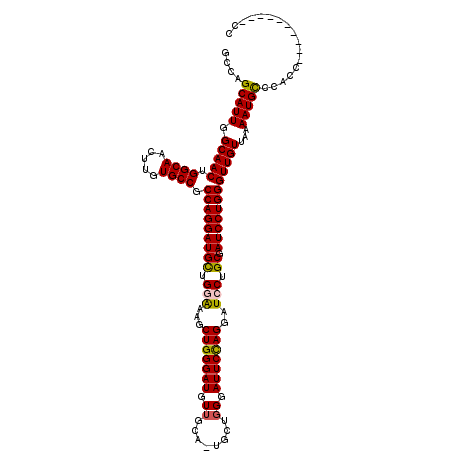

| Location | 7,204,272 – 7,204,391 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.21 |
| Mean single sequence MFE | -37.82 |
| Consensus MFE | -32.33 |
| Energy contribution | -32.83 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7204272 119 + 23771897 AGGAUACUAGAAUCCCAGCAUUGCAACAUCCCAGCUUUCCAACAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGCGACUAAUAGUGCUGAGCCCGGAGAGGAC-AAAAAAUAUCC .(((((..........(((..............))).(((....(((.((((((((.((((((((((.......))))))))))....)))))..))).)))..))).-......))))) ( -37.14) >DroSec_CAF1 17657 120 + 1 AGGAUCCUAGAAUCCCAGCAGUGCAACAUCCCAGCUUUCCAGCAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGCGACUAAUAGUGCUGAGCCCGGAGAGGACGAAAAAAUAUCC ....((((....((((((((.((.....(((((((......(((..((((((((....)))))))).)))....))))).))....)).))))).....))).))))............. ( -38.30) >DroEre_CAF1 18106 111 + 1 AGCAUCCUGGAAUCCCA--------ACAUCCCAGCUUCCCAGCAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGCGACUAAUAGUGCUGAGCCCGGAGAGCAC-AAAAAAUAUCC .((....(((....)))--------...(((..(((...((((((...((((((....))))))((((((((....))))))))....)))))))))..)))..))..-........... ( -37.30) >DroYak_CAF1 15261 119 + 1 UGGAUCCUGGAAUCCCAACAACCCAACAUCCCAGCUUCACAGCAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGCGACUAAUAGUGCUGAGCCCGGAGAGCAC-AAAAAAUAUCC ((((..((((....................))))..)).))((.(((.((((((((.((((((((((.......))))))))))....)))))..))).)))..))..-........... ( -38.55) >consensus AGGAUCCUAGAAUCCCAGCA_UGCAACAUCCCAGCUUCCCAGCAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGCGACUAAUAGUGCUGAGCCCGGAGAGCAC_AAAAAAUAUCC .((((......)))).............(((..(((...((((((...((((((....))))))((((((((....))))))))....)))))))))..))).................. (-32.33 = -32.83 + 0.50)



| Location | 7,204,312 – 7,204,431 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -45.68 |
| Consensus MFE | -40.88 |
| Energy contribution | -41.14 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7204312 119 + 23771897 AACAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGCGACUAAUAGUGCUGAGCCCGGAGAGGAC-AAAAAAUAUCCUUGCUGCCAGCGGCGUUUGUUUGCCUUGCCUCUUCAUUUU ........((((((((.((((((((((.......))))))))))....)))))..))).(((((((.(-((.(((((....(((((....)))))..)))))...))))))))))..... ( -45.90) >DroSec_CAF1 17697 120 + 1 AGCAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGCGACUAAUAGUGCUGAGCCCGGAGAGGACGAAAAAAUAUCCUUGCUGCCAGCGGCGUUUGUUUGCCUUGCCUCUUCAUUUU ........((((((((.((((((((((.......))))))))))....)))))..))).(((((((.(((..(((((....(((((....)))))..)))))...))))))))))..... ( -46.00) >DroEre_CAF1 18138 119 + 1 AGCAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGCGACUAAUAGUGCUGAGCCCGGAGAGCAC-AAAAAAUAUCCUUGCUGCCAGCGGCGUUUGUUUGCCUUGCCUCUUCAUUUU .(((..(((((((((..((((((((((.......))))))))))....(((((..........)))))-............))))))))).((((......)))).)))........... ( -45.40) >DroYak_CAF1 15301 119 + 1 AGCAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGCGACUAAUAGUGCUGAGCCCGGAGAGCAC-AAAAAAUAUCCUUGCUGCCAGCGGCGUUUGUUUGCCUUGCCCCUUCAUGUU .(((..(((((((((..((((((((((.......))))))))))....(((((..........)))))-............))))))))).((((......)))).)))........... ( -45.40) >consensus AGCAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGCGACUAAUAGUGCUGAGCCCGGAGAGCAC_AAAAAAUAUCCUUGCUGCCAGCGGCGUUUGUUUGCCUUGCCUCUUCAUUUU .(((..((((((((((.((((((((((.......))))))))))....))))).(((..(((...............)))..)))))))).((((......)))).)))........... (-40.88 = -41.14 + 0.25)

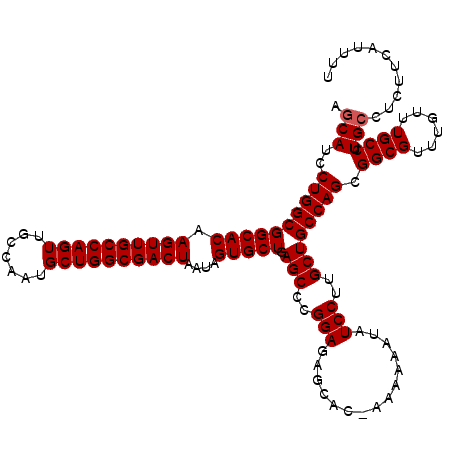
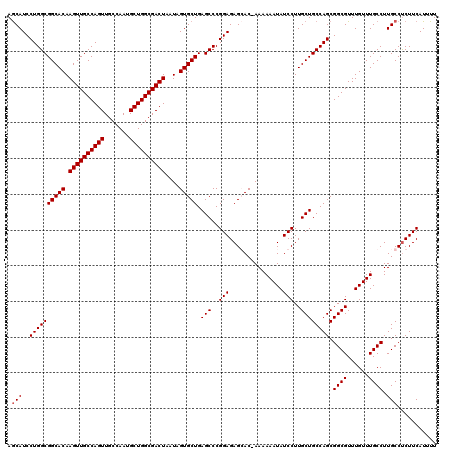
| Location | 7,204,431 – 7,204,551 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.47 |
| Mean single sequence MFE | -41.65 |
| Consensus MFE | -39.42 |
| Energy contribution | -39.42 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7204431 120 + 23771897 UCCCUCAAUUUAUACAUUUGCGCAUAAAAAGCAAGUGUAAAAGUUUUGCAUAUUCAUGGCGCUUUAAAUGUCGUCGCCAAGCCACGAGGAUGGCGACGUCUGAAGUGCAGUGGCAGCCAC ......(((((.(((((((((.........))))))))).)))))((((.....(((.((((((((...(.((((((((..((....)).)))))))).))))))))).))))))).... ( -41.10) >DroSec_CAF1 17817 120 + 1 UCCCUCAAUUUAUACAUUUGCGCAUAAAAAGCAAGUGUAAAAGUUUUGCAUAUUCAUGGCGCUUUAAAUGUCGUCGCCAAGCCACGAGGAUGGCGACGUCUGAAGUGCAGUGGCAGCCAC ......(((((.(((((((((.........))))))))).)))))((((.....(((.((((((((...(.((((((((..((....)).)))))))).))))))))).))))))).... ( -41.10) >DroEre_CAF1 18257 120 + 1 UCCCUCAAUUUAUACAUUUGCGCAUAAAAAGCAAGUGUAAAAGUUUUGCAUAUUCAUGGCGCUUUAAAUGUCGUCGCCAAGCCACGAGGAUGGCGACGUCUGAAGUGCAGAUGCAGCCAC ......(((((.(((((((((.........))))))))).)))))((((((.......((((((((...(.((((((((..((....)).)))))))).)))))))))..)))))).... ( -42.50) >DroYak_CAF1 15420 120 + 1 UCCCUCAAUUUAUACAUUUGCGCAUAAAAAGCAAGUGUAAAAGUUUUGCAUAUUCAUGGCGCUUUAAAUGUCGUCGCCAAGCCACGAGGAUGGCGACGUCUGAAGUGCGGAUGCAGCCAC ......(((((.(((((((((.........))))))))).)))))((((((.....(.((((((((...(.((((((((..((....)).)))))))).))))))))).))))))).... ( -41.90) >consensus UCCCUCAAUUUAUACAUUUGCGCAUAAAAAGCAAGUGUAAAAGUUUUGCAUAUUCAUGGCGCUUUAAAUGUCGUCGCCAAGCCACGAGGAUGGCGACGUCUGAAGUGCAGAGGCAGCCAC ......(((((.(((((((((.........))))))))).)))))...........((((((((((...(.((((((((..((....)).)))))))).)))))))((....)).)))). (-39.42 = -39.42 + 0.00)

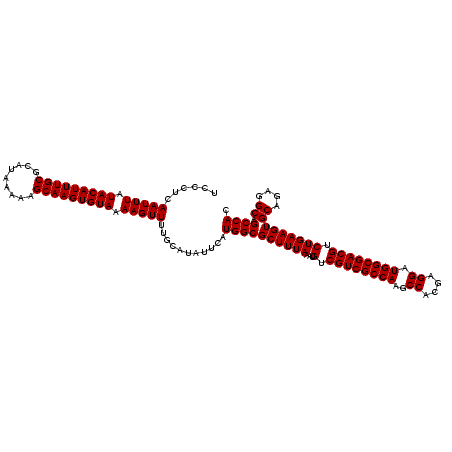

| Location | 7,204,431 – 7,204,551 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.47 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -39.91 |
| Energy contribution | -39.73 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7204431 120 - 23771897 GUGGCUGCCACUGCACUUCAGACGUCGCCAUCCUCGUGGCUUGGCGACGACAUUUAAAGCGCCAUGAAUAUGCAAAACUUUUACACUUGCUUUUUAUGCGCAAAUGUAUAAAUUGAGGGA ((((...))))....((((((.((((((((.((....))..))))))))((((((...((((.(((((...((((...........))))..))))))))))))))).....)))))).. ( -40.70) >DroSec_CAF1 17817 120 - 1 GUGGCUGCCACUGCACUUCAGACGUCGCCAUCCUCGUGGCUUGGCGACGACAUUUAAAGCGCCAUGAAUAUGCAAAACUUUUACACUUGCUUUUUAUGCGCAAAUGUAUAAAUUGAGGGA ((((...))))....((((((.((((((((.((....))..))))))))((((((...((((.(((((...((((...........))))..))))))))))))))).....)))))).. ( -40.70) >DroEre_CAF1 18257 120 - 1 GUGGCUGCAUCUGCACUUCAGACGUCGCCAUCCUCGUGGCUUGGCGACGACAUUUAAAGCGCCAUGAAUAUGCAAAACUUUUACACUUGCUUUUUAUGCGCAAAUGUAUAAAUUGAGGGA .....(((....)))((((((.((((((((.((....))..))))))))((((((...((((.(((((...((((...........))))..))))))))))))))).....)))))).. ( -39.40) >DroYak_CAF1 15420 120 - 1 GUGGCUGCAUCCGCACUUCAGACGUCGCCAUCCUCGUGGCUUGGCGACGACAUUUAAAGCGCCAUGAAUAUGCAAAACUUUUACACUUGCUUUUUAUGCGCAAAUGUAUAAAUUGAGGGA ((((......)))).((((((.((((((((.((....))..))))))))((((((...((((.(((((...((((...........))))..))))))))))))))).....)))))).. ( -41.60) >consensus GUGGCUGCAACUGCACUUCAGACGUCGCCAUCCUCGUGGCUUGGCGACGACAUUUAAAGCGCCAUGAAUAUGCAAAACUUUUACACUUGCUUUUUAUGCGCAAAUGUAUAAAUUGAGGGA ((((......)))).((((((.((((((((.((....))..))))))))((((((...((((.(((((...((((...........))))..))))))))))))))).....)))))).. (-39.91 = -39.73 + -0.19)

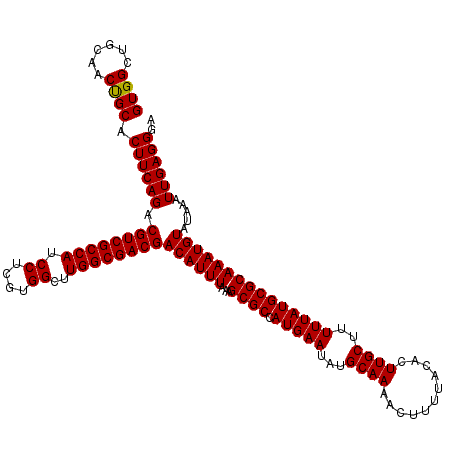
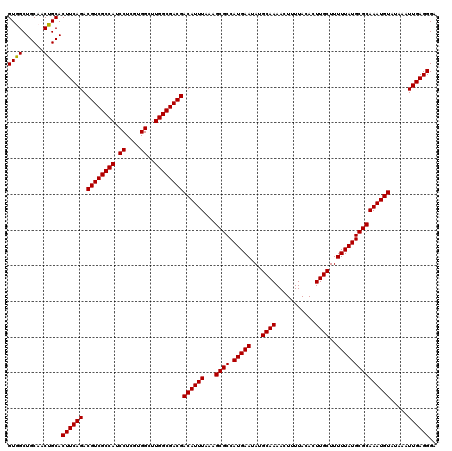
| Location | 7,204,471 – 7,204,591 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.47 |
| Mean single sequence MFE | -42.17 |
| Consensus MFE | -41.24 |
| Energy contribution | -42.05 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.38 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7204471 120 + 23771897 AAGUUUUGCAUAUUCAUGGCGCUUUAAAUGUCGUCGCCAAGCCACGAGGAUGGCGACGUCUGAAGUGCAGUGGCAGCCACAAAGGCAAAAAGCAAUGCUGCAACUAGCAUAAAAAACAAU ..(((((((....((((.((((((((...(.((((((((..((....)).)))))))).))))))))).))))..(((.....))).....)).((((((....))))))..)))))... ( -43.10) >DroSec_CAF1 17857 120 + 1 AAGUUUUGCAUAUUCAUGGCGCUUUAAAUGUCGUCGCCAAGCCACGAGGAUGGCGACGUCUGAAGUGCAGUGGCAGCCACAAAGGCAAAAAGCAAUGCUGCAACUAGCAUAAAAAACAAU ..(((((((....((((.((((((((...(.((((((((..((....)).)))))))).))))))))).))))..(((.....))).....)).((((((....))))))..)))))... ( -43.10) >DroEre_CAF1 18297 120 + 1 AAGUUUUGCAUAUUCAUGGCGCUUUAAAUGUCGUCGCCAAGCCACGAGGAUGGCGACGUCUGAAGUGCAGAUGCAGCCACAAAGGCAAAAAGCAAUGCUGCAACUAGCAUAAAAAACAAU ..(((((((((.......((((((((...(.((((((((..((....)).)))))))).)))))))))..)))).(((.....)))..))))).((((((....)))))).......... ( -41.50) >DroYak_CAF1 15460 120 + 1 AAGUUUUGCAUAUUCAUGGCGCUUUAAAUGUCGUCGCCAAGCCACGAGGAUGGCGACGUCUGAAGUGCGGAUGCAGCCACAAAGGCAAAAAGCAAUGCUGCAACUAGCAUAAAAAACAAU ..(((((((.(((((...((((((((...(.((((((((..((....)).)))))))).))))))))))))))..(((.....))).....)).((((((....))))))..)))))... ( -41.00) >consensus AAGUUUUGCAUAUUCAUGGCGCUUUAAAUGUCGUCGCCAAGCCACGAGGAUGGCGACGUCUGAAGUGCAGAGGCAGCCACAAAGGCAAAAAGCAAUGCUGCAACUAGCAUAAAAAACAAU ..(((((((.........((((((((...(.((((((((..((....)).)))))))).)))))))))(((((((((.......((.....))...))))))))).)))....))))... (-41.24 = -42.05 + 0.81)

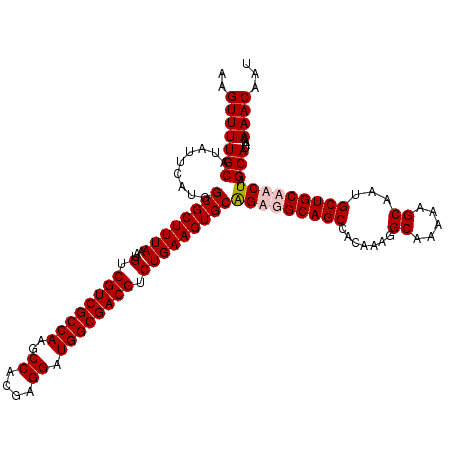
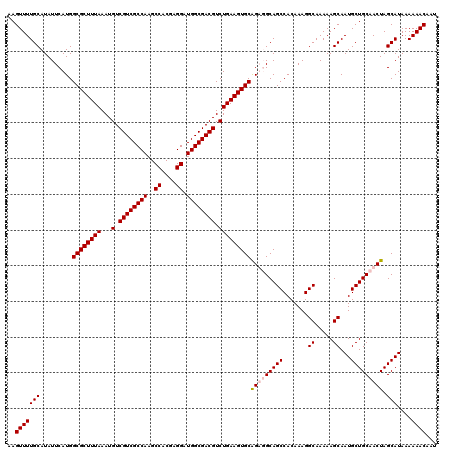
| Location | 7,204,471 – 7,204,591 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.47 |
| Mean single sequence MFE | -37.48 |
| Consensus MFE | -36.33 |
| Energy contribution | -37.58 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7204471 120 - 23771897 AUUGUUUUUUAUGCUAGUUGCAGCAUUGCUUUUUGCCUUUGUGGCUGCCACUGCACUUCAGACGUCGCCAUCCUCGUGGCUUGGCGACGACAUUUAAAGCGCCAUGAAUAUGCAAAACUU .((((..((((((..(((.(((((...((.....)).......))))).)))((.(((.(((((((((((.((....))..))))))))...))).))).))))))))...))))..... ( -37.20) >DroSec_CAF1 17857 120 - 1 AUUGUUUUUUAUGCUAGUUGCAGCAUUGCUUUUUGCCUUUGUGGCUGCCACUGCACUUCAGACGUCGCCAUCCUCGUGGCUUGGCGACGACAUUUAAAGCGCCAUGAAUAUGCAAAACUU .((((..((((((..(((.(((((...((.....)).......))))).)))((.(((.(((((((((((.((....))..))))))))...))).))).))))))))...))))..... ( -37.20) >DroEre_CAF1 18297 120 - 1 AUUGUUUUUUAUGCUAGUUGCAGCAUUGCUUUUUGCCUUUGUGGCUGCAUCUGCACUUCAGACGUCGCCAUCCUCGUGGCUUGGCGACGACAUUUAAAGCGCCAUGAAUAUGCAAAACUU ..........(((((......)))))....((((((.((..((((.((.((((.....))))((((((((.((....))..)))))))).........))))))..))...))))))... ( -39.10) >DroYak_CAF1 15460 120 - 1 AUUGUUUUUUAUGCUAGUUGCAGCAUUGCUUUUUGCCUUUGUGGCUGCAUCCGCACUUCAGACGUCGCCAUCCUCGUGGCUUGGCGACGACAUUUAAAGCGCCAUGAAUAUGCAAAACUU ..........(((((......)))))....((((((.((..(((((((....))).......((((((((.((....))..))))))))...........))))..))...))))))... ( -36.40) >consensus AUUGUUUUUUAUGCUAGUUGCAGCAUUGCUUUUUGCCUUUGUGGCUGCAACUGCACUUCAGACGUCGCCAUCCUCGUGGCUUGGCGACGACAUUUAAAGCGCCAUGAAUAUGCAAAACUU .((((..((((((..(((((((((...((.....)).......)))))))))((.(((.(((((((((((.((....))..))))))))...))).))).))))))))...))))..... (-36.33 = -37.58 + 1.25)

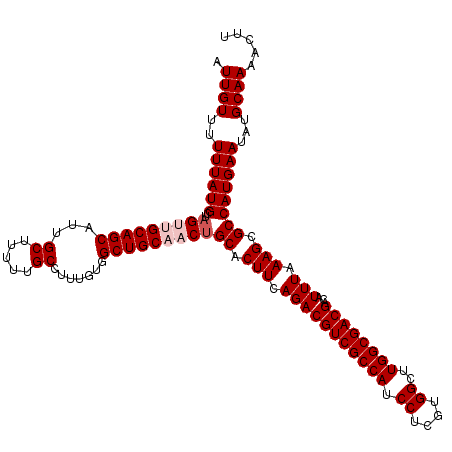

| Location | 7,204,511 – 7,204,631 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -34.06 |
| Energy contribution | -34.88 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7204511 120 + 23771897 GCCACGAGGAUGGCGACGUCUGAAGUGCAGUGGCAGCCACAAAGGCAAAAAGCAAUGCUGCAACUAGCAUAAAAAACAAUAACCAGCUGCUGAAGUUAGUUAUGUUAGAUGAUACCCUGC ((((......))))..(((((((.(((((((.(((((.......((.....))...))))).))).))))........(((((.((((.....)))).))))).)))))))......... ( -34.00) >DroSec_CAF1 17897 120 + 1 GCCACGAGGAUGGCGACGUCUGAAGUGCAGUGGCAGCCACAAAGGCAAAAAGCAAUGCUGCAACUAGCAUAAAAAACAAUAACCAGCUGCUGAAGUUAGUUAUGUUAGAUGAUACCCUAA ((((......))))..(((((((.(((((((.(((((.......((.....))...))))).))).))))........(((((.((((.....)))).))))).)))))))......... ( -34.00) >DroEre_CAF1 18337 120 + 1 GCCACGAGGAUGGCGACGUCUGAAGUGCAGAUGCAGCCACAAAGGCAAAAAGCAAUGCUGCAACUAGCAUAAAAAACAAUAACCAGCUGCUGAAGUUAGUUAUGUUAGAUGAUGCCCUAA .((....))..((((.(((((((.((((((.((((((.......((.....))...)))))).)).))))........(((((.((((.....)))).))))).))))))).)))).... ( -35.40) >DroYak_CAF1 15500 120 + 1 GCCACGAGGAUGGCGACGUCUGAAGUGCGGAUGCAGCCACAAAGGCAAAAAGCAAUGCUGCAACUAGCAUAAAAAACAAUAACCAGCUGCUGAAGUUAGUUAUGUUAGAUGAUGCCCUAA .((....))..((((.(((((((.((((((.((((((.......((.....))...)))))).)).))))........(((((.((((.....)))).))))).))))))).)))).... ( -34.70) >consensus GCCACGAGGAUGGCGACGUCUGAAGUGCAGAGGCAGCCACAAAGGCAAAAAGCAAUGCUGCAACUAGCAUAAAAAACAAUAACCAGCUGCUGAAGUUAGUUAUGUUAGAUGAUACCCUAA ((((......))))..(((((((.(((((((((((((.......((.....))...))))))))).))))........(((((.((((.....)))).))))).)))))))......... (-34.06 = -34.88 + 0.81)


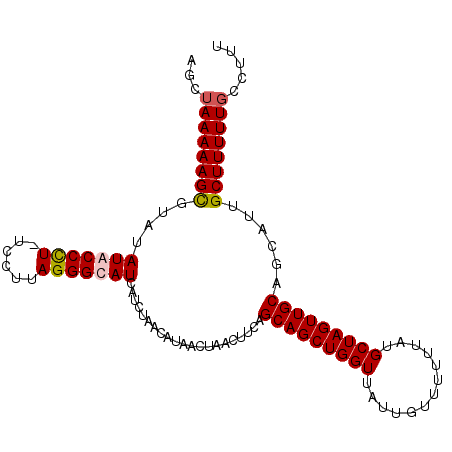
| Location | 7,204,551 – 7,204,657 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -24.60 |
| Energy contribution | -24.85 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7204551 106 + 23771897 AAAGGCAAAAAGCAAUGCUGCAACUAGCAUAAAAAACAAUAACCAGCUGCUGAAGUUAGUUAUGUUAGAUGAUACCCUGCAAAAAGGGUAUAUACACUUUUUAACU ..((..((((((..((((((....))))))....((((.((((.((((.....)))).)))))))).....(((((((......))))))).....))))))..)) ( -25.40) >DroSec_CAF1 17937 106 + 1 AAAGGCAAAAAGCAAUGCUGCAACUAGCAUAAAAAACAAUAACCAGCUGCUGAAGUUAGUUAUGUUAGAUGAUACCCUAAGGAGAGGGUAUAUACGCUUUUUAGCU ...((((((((((.((((((....))))))....((((.((((.((((.....)))).)))))))).....(((((((......)))))))....))))))).))) ( -30.90) >DroEre_CAF1 18377 106 + 1 AAAGGCAAAAAGCAAUGCUGCAACUAGCAUAAAAAACAAUAACCAGCUGCUGAAGUUAGUUAUGUUAGAUGAUGCCCUAAGGAUAGGGUAUAGGCGCUUUUUAGAU ......(((((((.((((((....))))))....((((.((((.((((.....)))).)))))))).(...((((((((....))))))))...)))))))).... ( -29.40) >DroYak_CAF1 15540 105 + 1 AAAGGCAAAAAGCAAUGCUGCAACUAGCAUAAAAAACAAUAACCAGCUGCUGAAGUUAGUUAUGUUAGAUGAUGCCCUAAGAA-AAGGUAUAUACGCUUUUUCGAU ...((((.......((((((....))))))....((((.((((.((((.....)))).))))))))......))))....(((-(((((......))))))))... ( -25.00) >consensus AAAGGCAAAAAGCAAUGCUGCAACUAGCAUAAAAAACAAUAACCAGCUGCUGAAGUUAGUUAUGUUAGAUGAUACCCUAAGAA_AGGGUAUAUACGCUUUUUAGAU ......(((((((.((((((....))))))....((((.((((.((((.....)))).)))))))).....(((((((......)))))))....))))))).... (-24.60 = -24.85 + 0.25)



| Location | 7,204,551 – 7,204,657 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -23.65 |
| Energy contribution | -24.02 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7204551 106 - 23771897 AGUUAAAAAGUGUAUAUACCCUUUUUGCAGGGUAUCAUCUAACAUAACUAACUUCAGCAGCUGGUUAUUGUUUUUUAUGCUAGUUGCAGCAUUGCUUUUUGCCUUU ((.((((((((....(((((((......))))))).....................(((((((((.............)))))))))......)))))))).)).. ( -25.62) >DroSec_CAF1 17937 106 - 1 AGCUAAAAAGCGUAUAUACCCUCUCCUUAGGGUAUCAUCUAACAUAACUAACUUCAGCAGCUGGUUAUUGUUUUUUAUGCUAGUUGCAGCAUUGCUUUUUGCCUUU .((.((((((((...(((((((......))))))).....................(((((((((.............))))))))).....)))))))))).... ( -28.82) >DroEre_CAF1 18377 106 - 1 AUCUAAAAAGCGCCUAUACCCUAUCCUUAGGGCAUCAUCUAACAUAACUAACUUCAGCAGCUGGUUAUUGUUUUUUAUGCUAGUUGCAGCAUUGCUUUUUGCCUUU ...((((((((.......(((((....)))))........................(((((((((.............)))))))))......))))))))..... ( -25.92) >DroYak_CAF1 15540 105 - 1 AUCGAAAAAGCGUAUAUACCUU-UUCUUAGGGCAUCAUCUAACAUAACUAACUUCAGCAGCUGGUUAUUGUUUUUUAUGCUAGUUGCAGCAUUGCUUUUUGCCUUU ....((((((((......(((.-.....))).........................(((((((((.............))))))))).....))))))))...... ( -21.62) >consensus AGCUAAAAAGCGUAUAUACCCU_UCCUUAGGGCAUCAUCUAACAUAACUAACUUCAGCAGCUGGUUAUUGUUUUUUAUGCUAGUUGCAGCAUUGCUUUUUGCCUUU ...((((((((....(((((((......))))))).....................(((((((((.............)))))))))......))))))))..... (-23.65 = -24.02 + 0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:53 2006