
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,202,724 – 7,203,001 |
| Length | 277 |
| Max. P | 0.634480 |

| Location | 7,202,724 – 7,202,844 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.35 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -27.40 |
| Energy contribution | -27.46 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
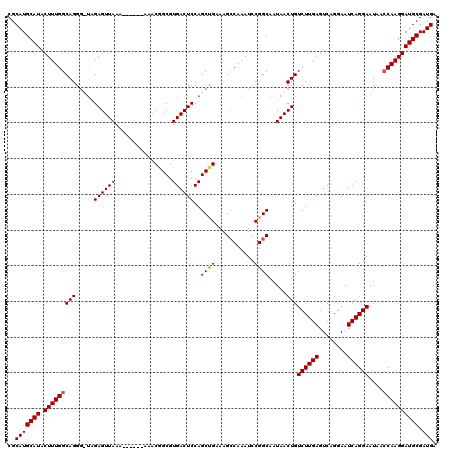
>3L_DroMel_CAF1 7202724 120 - 23771897 CGCAUGCAUACUUUGGCAGGGUUAGAGUUAAAAAAAAAAAACGGCGUGACUCCAGCUGGAAGCCAAAUCCGGCAAUAACUGUCUUGAGUCGGGAAUCAGGAAUCACCAAGGAUGCGAUGA ..(((((((.((((((...(((((((((((................))))))..((((((.......))))))..))))).((((((........))))))....)))))))))).))). ( -34.19) >DroSec_CAF1 16140 116 - 1 CGCAUGCAUACUUUGGCAGGGUUAGAGUUAAAA----AAAACGGCGUGACUCCAGCUGUAAGGCAAAUCCGGCAAUAACUGUCUUGAGUCGGGAAUCAGGAAUCACCAAGGAUGCGAUGA (((((.(..((((((((...)))))))).....----......(.((((.(((.(((....)))...((((((..............)))))).....))).)))))..).))))).... ( -30.94) >DroEre_CAF1 16597 113 - 1 CGCAUGCAUACUUUGGCAGGA-CAGAGUUAAA------AAGCGGCGUGACUCCAGCCGAAAGCCAAAUCCGGCAAUAACUGUCUUGAGUCAGGAAUCAGGAAUACCCAAGGAUGCGAUGA ..(((((((.(((((((((((-(((.......------...((((.((....))))))...(((......))).....))))))))..((........)).....)))))))))).))). ( -34.30) >DroYak_CAF1 13667 113 - 1 CGCAUGCAUACUUUGGCAGGG-UGGAGUUAAA------AAGCCGCGUGACUCCAGCUGAAAGCCAAAUCCGGCAAUAACUGUCUUGAGUCAGGAAUCAGGAAUAAGCAAGGAUGCGAUGU (((((((....((((((.((.-((((((((..------........)))))))).))....))))))....)).....((.((((((........))))))...)).....))))).... ( -34.50) >consensus CGCAUGCAUACUUUGGCAGGG_UAGAGUUAAA______AAACGGCGUGACUCCAGCUGAAAGCCAAAUCCGGCAAUAACUGUCUUGAGUCAGGAAUCAGGAAUAACCAAGGAUGCGAUGA ..(((((((.(((((((((.....((((((................))))))..((((...........)))).....)))((((((........))))))....)))))))))).))). (-27.40 = -27.46 + 0.06)


| Location | 7,202,844 – 7,202,961 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.15 |
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -16.41 |
| Energy contribution | -18.22 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.515955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
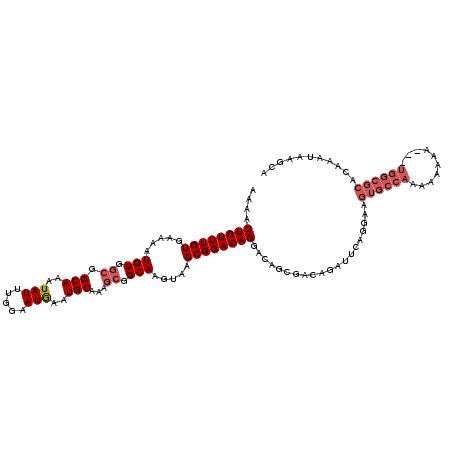
>3L_DroMel_CAF1 7202844 117 + 23771897 AAAAUUGUUUGUGAAAAUGUACCGGCAAAUAUUUGGAAUGAAUGCAAAGCGGCAAGUAAGCGGACAGGACAGCGACAGAUUCAGGAUGUGCCAAAAAAAA---UGGCGCACAAAUAAGCA ....(((((((((((..(((((..((.....((((.(.....).))))...))..))..((..........)).)))..))).....((((((.......---))))))))))))))... ( -23.10) >DroSec_CAF1 16256 109 + 1 AAAAUUGUUUGUGAAAAUGUGGCGGCAAAUAUUUGGAAUGAAUGCAAAGCGGCAAGUAAGCGGACAGGACAGCGACAAAUUCAGGAUGUGCAAAAAAAAA-----------AAAAAAGCA ....((((((((.....(((.((.(((..(((.....)))..)))...)).))).....))))))))....((.(((.........))))).........-----------......... ( -18.20) >DroEre_CAF1 16710 116 + 1 AAAAUUGUUUGUGAAAAUGUGGCGGCAAAUAUUUGGAAUGAAUGCAAAGCGGCAAGUAAGCGGACAGGACAGCGACAGAUUCAGGAAGUGCCAAAAAAA----UGGCGCACAAAUAAGAA ....(((((((((((..(((.((.(((..(((.....)))..)))...((.((......)).).)......)).)))..))).....((((((......----))))))))))))))... ( -28.50) >DroYak_CAF1 13780 120 + 1 AAAAUUGUUUGUGCAAAUGUGGCGGCAAAUAUUUGAAAUAAAUGCAAAACGGCAAGUAAGCGGACAGGACAGCGACAGAUUCAGGAAGUGCCAAAAAAAAAAUUGGCGCACAAAUAAGCA ....((((((((((...(((.((.(((..(((.....)))..)))....(.((......)).)........)).)))............(((((........)))))))))))))))... ( -29.60) >consensus AAAAUUGUUUGUGAAAAUGUGGCGGCAAAUAUUUGGAAUGAAUGCAAAGCGGCAAGUAAGCGGACAGGACAGCGACAGAUUCAGGAAGUGCCAAAAAAAA___UGGCGCACAAAUAAGCA ....((((((((.....(((.((.(((..(((.....)))..)))...)).))).....))))))))....................((((((..........))))))........... (-16.41 = -18.22 + 1.81)


| Location | 7,202,884 – 7,203,001 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.18 |
| Mean single sequence MFE | -25.77 |
| Consensus MFE | -17.94 |
| Energy contribution | -19.25 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
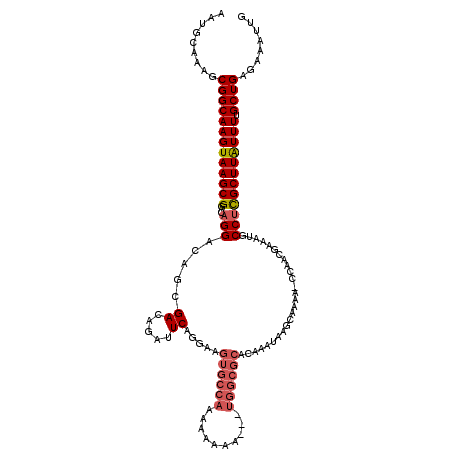
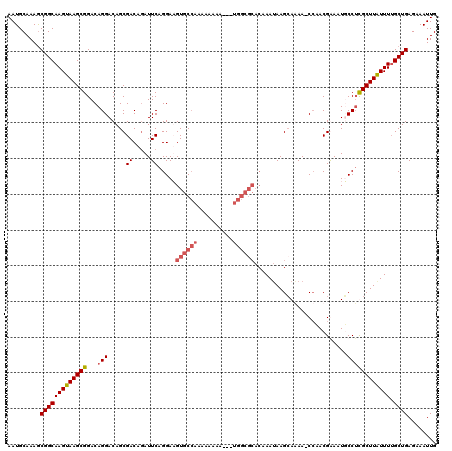
>3L_DroMel_CAF1 7202884 117 + 23771897 AAUGCAAAGCGGCAAGUAAGCGGACAGGACAGCGACAGAUUCAGGAUGUGCCAAAAAAAA---UGGCGCACAAAUAAGCAAAAACCCACGGAAUGCCAUGCUUAUUUUGCUGAGAAAUUG .........(((((((((((((....((.((((((.....))....(((((((.......---))))))).......)).....((...))..)))).)))))).)))))))........ ( -26.90) >DroSec_CAF1 16296 109 + 1 AAUGCAAAGCGGCAAGUAAGCGGACAGGACAGCGACAAAUUCAGGAUGUGCAAAAAAAAA-----------AAAAAAGCAAAACCAAACGGAAUGCCUUGCUUAUUUGGCUGAGAAAUUG .........(((((((((((((...(((.((..((.....)).((...(((.........-----------......)))...))........)))))))))))))).))))........ ( -20.06) >DroEre_CAF1 16750 115 + 1 AAUGCAAAGCGGCAAGUAAGCGGACAGGACAGCGACAGAUUCAGGAAGUGCCAAAAAAA----UGGCGCACAAAUAAGAAGAA-CCAACGAAAUUCCUCGCUUGUUUUGCUGAGAAAUUG ..(((......)))......(((.(((((((((((..((((..(...((((((......----))))))........(.....-.)..)..))))..)))).))))))))))........ ( -27.20) >DroYak_CAF1 13820 119 + 1 AAUGCAAAACGGCAAGUAAGCGGACAGGACAGCGACAGAUUCAGGAAGUGCCAAAAAAAAAAUUGGCGCACAAAUAAGCAGAA-CCAACGAAAUUCCUCGCUUAUUUUGCUGAGAAAUUG .........(((((((((((((...((((..((((.....)).....(((((((........)))))))........)).(..-....).....)))))))))).)))))))........ ( -28.90) >consensus AAUGCAAAGCGGCAAGUAAGCGGACAGGACAGCGACAGAUUCAGGAAGUGCCAAAAAAAA___UGGCGCACAAAUAAGCAAAA_CCAACGAAAUGCCUCGCUUAUUUUGCUGAGAAAUUG .........(((((((((((((...(((.....((.....)).....((((((..........))))))..........................)))))))))))).))))........ (-17.94 = -19.25 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:43 2006