
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
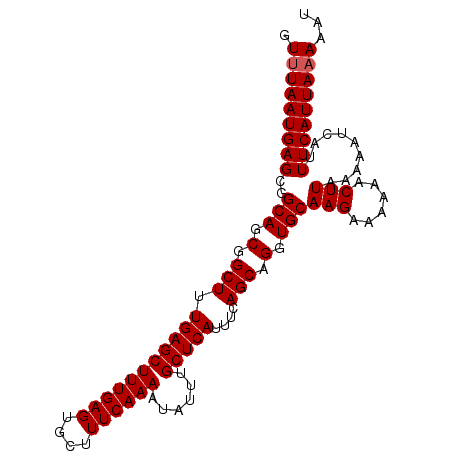
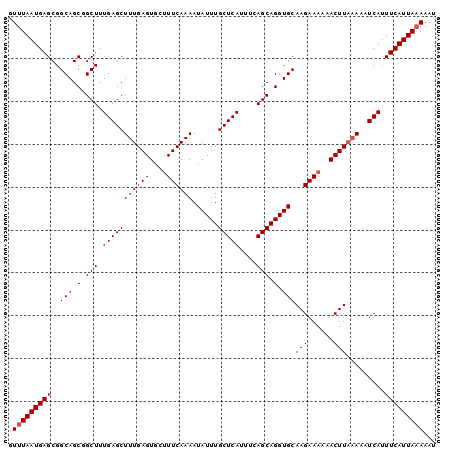
| Location | 7,200,409 – 7,200,549 |
| Length | 140 |
| Max. P | 0.920095 |

| Location | 7,200,409 – 7,200,509 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.68 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7200409 100 - 23771897 GUUUAAUGAGCGGCAGCGGCUUUGAGCUUUGAGUGCUUUCAAAAUAUUUGCUCAUUUCAGCAGGUGCAAGAAAAAACUUAAAAAUCAUUUCAUUACAAAU ...((((((((....)).....(((..(((((((..((((....((((((((......))))))))...))))..)))))))..)))..))))))..... ( -22.60) >DroSec_CAF1 13863 100 - 1 GUUUAAUGAGCGGCAGCGGCUUUGAGCUUUGAGUGCUUUCAAAAUAUUUGCUCAUUUCAGCAGGUGCAAGAAAAAACUUCAAAAUCAUUUCAUUAAAAAU .(((((((((..(((.(.(((.(((((((((((....))))))......)))))....))).).)))..(((.....)))........)))))))))... ( -23.20) >DroEre_CAF1 14314 100 - 1 GUUUAAUGAGCGGCAGCGGCUUUGAGCUUUGAGUGGCUUCAAAAUAUUUGCUCAUUUUAGCAGGUGCAAGAAAAAACUUACAAAUCAUUUCAUUAAAAAU .(((((((((..(((.(.(((.(((((((((((....))))))......)))))....))).).)))(((......))).........)))))))))... ( -22.20) >DroYak_CAF1 11337 100 - 1 GUUUAAUGAGCGGCAGCCGCUUUGAGCUUUGAGUGCUUUCAAAAUAUUUGCUCAUUUCAGCAGGUGCAAGAAAAAACUUAAAAAUCAUUUCAUUAAAAAU .((((((((((((...))))..(((..(((((((..((((....((((((((......))))))))...))))..)))))))..)))..))))))))... ( -27.60) >consensus GUUUAAUGAGCGGCAGCGGCUUUGAGCUUUGAGUGCUUUCAAAAUAUUUGCUCAUUUCAGCAGGUGCAAGAAAAAACUUAAAAAUCAUUUCAUUAAAAAU .(((((((((..(((.(.(((.(((((((((((....))))))......)))))....))).).)))(((......))).........)))))))))... (-21.42 = -21.68 + 0.25)

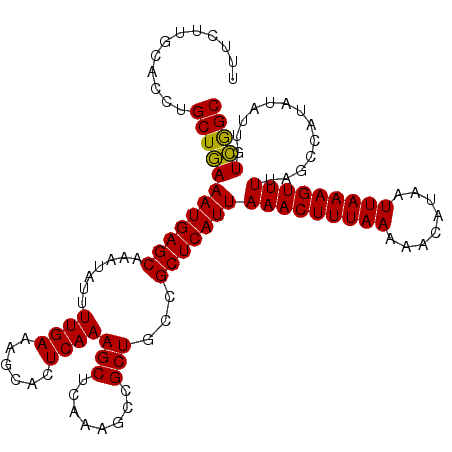
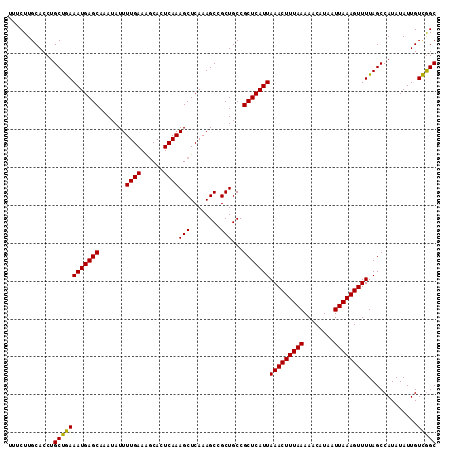
| Location | 7,200,436 – 7,200,549 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 97.35 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -22.86 |
| Energy contribution | -22.30 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7200436 113 + 23771897 UUUCUUGCACCUGCUGAAAUGAGCAAAUAUUUUGAAAGCACUCAAAGCUCAAAGCCGCUGCCGCUCAUUAAACUUUAAAAACAUAAUUAAAGUUUUAGCCAUAUAUUGUCAGC ............((((((((((((.......((((......))))(((........)))...)))))))(((((((((........))))))))).............))))) ( -23.20) >DroSec_CAF1 13890 113 + 1 UUUCUUGCACCUGCUGAAAUGAGCAAAUAUUUUGAAAGCACUCAAAGCUCAAAGCCGCUGCCGCUCAUUAAACUUUAAAAACAUAAUUAAAGUUUUAGCCAUAUAUUGUCGGC ............((((((((((((.......((((......))))(((........)))...)))))))(((((((((........))))))))).............))))) ( -22.50) >DroEre_CAF1 14341 113 + 1 UUUCUUGCACCUGCUAAAAUGAGCAAAUAUUUUGAAGCCACUCAAAGCUCAAAGCCGCUGCCGCUCAUUAAACUUUAAAAACAUAAUUAAAGUUUUAGCCAUAUAUUGUUGGC ......((....))...(((((((......(((((.((........)))))))((....)).)))))))(((((((((........)))))))))..((((........)))) ( -23.30) >DroYak_CAF1 11364 113 + 1 UUUCUUGCACCUGCUGAAAUGAGCAAAUAUUUUGAAAGCACUCAAAGCUCAAAGCGGCUGCCGCUCAUUAAACUUUAAAAACAUAAUUAAAGUUUUAGCCAUAUAUUGUCGGC ......(((.(((((....(((((......(((((......)))))))))).))))).))).(((....(((((((((........))))))))).))).............. ( -24.50) >consensus UUUCUUGCACCUGCUGAAAUGAGCAAAUAUUUUGAAAGCACUCAAAGCUCAAAGCCGCUGCCGCUCAUUAAACUUUAAAAACAUAAUUAAAGUUUUAGCCAUAUAUUGUCGGC ............((((((((((((.......((((......))))(((........)))...)))))))(((((((((........))))))))).............))))) (-22.86 = -22.30 + -0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:38 2006