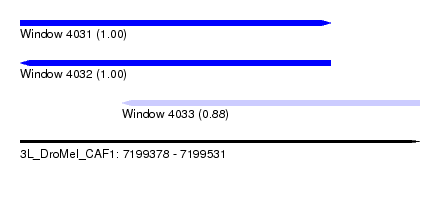
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,199,378 – 7,199,531 |
| Length | 153 |
| Max. P | 0.999785 |

| Location | 7,199,378 – 7,199,497 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.82 |
| Mean single sequence MFE | -35.24 |
| Consensus MFE | -31.68 |
| Energy contribution | -32.08 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.07 |
| SVM RNA-class probability | 0.999785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7199378 119 + 23771897 -UCUCUGGCUGCAGUGCAAUGCUUCUUCAUUCCUGGCAUUCGCACGUCACUCAGCAUCAAGCGGAAACAGUUAGCAACCAUCAUGGCUACCUCGCCAUCUUGCUUUCUCUCUCUCCCGCU -......((((..((((((((((...........)))))).))))......))))....(((((....((..(((((.....(((((......))))).)))))..)).......))))) ( -32.90) >DroSec_CAF1 12918 119 + 1 -UCUCUGGCUGCAGUGCAAUGCUUCUUCAUUCCUGGCAUUCGCACGUCACUCAGCAUCAAGCGGAAACAGUUAGCAACCAUCAUGGCUACCUCGCCAUCUUGCUCGCUCUCUCUCCCGCU -......((((..((((((((((...........)))))).))))......))))....(((((....((..(((...((..(((((......)))))..))...)))..))...))))) ( -34.60) >DroSim_CAF1 4872 119 + 1 -UCUCUGGCUGCAGUGCAAUGCUUCUUCAUUCCUGGCAUUCGCACGUCACUCAGCAUCAAGCGGAAACAGUUAGCAACCAUCAUGGCUACCUCGCCAUCUUGCUUGCUCUCUCUCCCGCU -......((((..((((((((((...........)))))).))))......))))....(((((....((..(((((.((..(((((......)))))..)).)))))..))...))))) ( -38.50) >DroEre_CAF1 13263 120 + 1 CUGGCUGGCUGCAGUGCAAUGCUUCUUCAUUCCUGGCAUUCGCACGUCACUCAGCAUCAAGCGGAAACAGUUAGCAACCAUCAUGGCUACCUCGCCAUCUUGCUUUCCCUCUCUCCCGCU ...((((..((..((((((((((...........)))))).))))..))..))))....(((((....((..(((((.....(((((......))))).)))))....)).....))))) ( -35.40) >DroYak_CAF1 10392 119 + 1 -UGGCUGGCUGCAGUGCAAUGCUUCUUCAUUCCUGGCAUUCGCACGUCACUCAGCAUCAAGCGGAAACAGUUAGCAACCAUCAUGGCUACCUCGCCAUAUUGCUCUCCCUCUCUCCCGCU -..((((..((..((((((((((...........)))))).))))..))..))))....(((((.....(..(((((.....(((((......))))).)))))..)........))))) ( -34.82) >consensus _UCUCUGGCUGCAGUGCAAUGCUUCUUCAUUCCUGGCAUUCGCACGUCACUCAGCAUCAAGCGGAAACAGUUAGCAACCAUCAUGGCUACCUCGCCAUCUUGCUUUCUCUCUCUCCCGCU .......((((..((((((((((...........)))))).))))......))))....(((((....((..(((((.....(((((......))))).)))))..)).......))))) (-31.68 = -32.08 + 0.40)



| Location | 7,199,378 – 7,199,497 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.82 |
| Mean single sequence MFE | -40.77 |
| Consensus MFE | -37.22 |
| Energy contribution | -36.74 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7199378 119 - 23771897 AGCGGGAGAGAGAGAAAGCAAGAUGGCGAGGUAGCCAUGAUGGUUGCUAACUGUUUCCGCUUGAUGCUGAGUGACGUGCGAAUGCCAGGAAUGAAGAAGCAUUGCACUGCAGCCAGAGA- ((((((((((..((.((.((..(((((......)))))..)).)).))..)).))))))))....((((((((..((((...(..((....)).)...))))..)))).))))......- ( -36.60) >DroSec_CAF1 12918 119 - 1 AGCGGGAGAGAGAGCGAGCAAGAUGGCGAGGUAGCCAUGAUGGUUGCUAACUGUUUCCGCUUGAUGCUGAGUGACGUGCGAAUGCCAGGAAUGAAGAAGCAUUGCACUGCAGCCAGAGA- ((((((((((..(((((.((..(((((......)))))..)).)))))..)).))))))))....((((((((..((((...(..((....)).)...))))..)))).))))......- ( -42.90) >DroSim_CAF1 4872 119 - 1 AGCGGGAGAGAGAGCAAGCAAGAUGGCGAGGUAGCCAUGAUGGUUGCUAACUGUUUCCGCUUGAUGCUGAGUGACGUGCGAAUGCCAGGAAUGAAGAAGCAUUGCACUGCAGCCAGAGA- ((((((((((..(((((.((..(((((......)))))..)).)))))..)).))))))))....((((((((..((((...(..((....)).)...))))..)))).))))......- ( -43.20) >DroEre_CAF1 13263 120 - 1 AGCGGGAGAGAGGGAAAGCAAGAUGGCGAGGUAGCCAUGAUGGUUGCUAACUGUUUCCGCUUGAUGCUGAGUGACGUGCGAAUGCCAGGAAUGAAGAAGCAUUGCACUGCAGCCAGCCAG ((((((((((..((.((.((..(((((......)))))..)).)).))..)).))))))))....((((..((..((((.(((((.............)))))))))..))..))))... ( -38.92) >DroYak_CAF1 10392 119 - 1 AGCGGGAGAGAGGGAGAGCAAUAUGGCGAGGUAGCCAUGAUGGUUGCUAACUGUUUCCGCUUGAUGCUGAGUGACGUGCGAAUGCCAGGAAUGAAGAAGCAUUGCACUGCAGCCAGCCA- ((((((((...((...(((((((((((......)))))....))))))..)).))))))))....((((..((..((((.(((((.............)))))))))..))..))))..- ( -42.22) >consensus AGCGGGAGAGAGAGAAAGCAAGAUGGCGAGGUAGCCAUGAUGGUUGCUAACUGUUUCCGCUUGAUGCUGAGUGACGUGCGAAUGCCAGGAAUGAAGAAGCAUUGCACUGCAGCCAGAGA_ ((((((((((..((.((.((..(((((......)))))..)).)).))..)).))))))))....((((((((..((((...(..((....)).)...))))..)))).))))....... (-37.22 = -36.74 + -0.48)


| Location | 7,199,417 – 7,199,531 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.24 |
| Mean single sequence MFE | -41.38 |
| Consensus MFE | -29.71 |
| Energy contribution | -29.75 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7199417 114 - 23771897 GGACACACACUCGCACGCUC-GCUGGCGGA-----AAGAGAGCGGGAGAGAGAGAAAGCAAGAUGGCGAGGUAGCCAUGAUGGUUGCUAACUGUUUCCGCUUGAUGCUGAGUGACGUGCG ...........((((((.((-(((((((..-----....(((((((((((..((.((.((..(((((......)))))..)).)).))..)).)))))))))..)))).))))))))))) ( -44.00) >DroSec_CAF1 12957 109 - 1 GG-CACAUA----CACACUC-GCAGGCGGA-----AAGAGAGCGGGAGAGAGAGCGAGCAAGAUGGCGAGGUAGCCAUGAUGGUUGCUAACUGUUUCCGCUUGAUGCUGAGUGACGUGCG .(-(((...----..(((((-(((((((((-----((.((..(....)....(((((.((..(((((......)))))..)).)))))..)).))))))))))....))))))..)))). ( -45.30) >DroSim_CAF1 4911 110 - 1 GGACACAUA----CACACUC-GCAGGCGGA-----AAGAGAGCGGGAGAGAGAGCAAGCAAGAUGGCGAGGUAGCCAUGAUGGUUGCUAACUGUUUCCGCUUGAUGCUGAGUGACGUGCG .........----..(((((-((.((((..-----....(((((((((((..(((((.((..(((((......)))))..)).)))))..)).)))))))))..))))..)))).))).. ( -42.00) >DroEre_CAF1 13303 117 - 1 GGACACACACACACACGC---ACGCUUGCAGGCGGAAGAGAGCGGGAGAGAGGGAAAGCAAGAUGGCGAGGUAGCCAUGAUGGUUGCUAACUGUUUCCGCUUGAUGCUGAGUGACGUGCG ...............(((---(((.(..(.((((.....(((((((((((..((.((.((..(((((......)))))..)).)).))..)).)))))))))..))))..)..))))))) ( -41.50) >DroYak_CAF1 10431 115 - 1 GGACGAACACACACACACUUAACACAUGAA-----AAGAGAGCGGGAGAGAGGGAGAGCAAUAUGGCGAGGUAGCCAUGAUGGUUGCUAACUGUUUCCGCUUGAUGCUGAGUGACGUGCG ...........(((.((((((.((..(...-----.)..(((((((((...((...(((((((((((......)))))....))))))..)).)))))))))..)).))))))..))).. ( -34.10) >consensus GGACACACAC_C_CACACUC_GCAGGCGGA_____AAGAGAGCGGGAGAGAGAGAAAGCAAGAUGGCGAGGUAGCCAUGAUGGUUGCUAACUGUUUCCGCUUGAUGCUGAGUGACGUGCG ...............(((((.(((...............(((((((((((..((.((.((..(((((......)))))..)).)).))..)).)))))))))..))).)))))....... (-29.71 = -29.75 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:36 2006