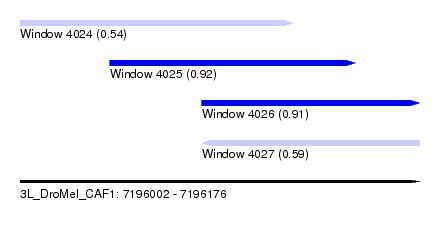
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,196,002 – 7,196,176 |
| Length | 174 |
| Max. P | 0.922595 |

| Location | 7,196,002 – 7,196,121 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.48 |
| Mean single sequence MFE | -40.18 |
| Consensus MFE | -36.28 |
| Energy contribution | -36.68 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7196002 119 + 23771897 UCCUGUCUU-CUGUAUCCUGUCCCGGAGCUGUCCUGUUCUCCUAGCUGGGCGGACGUCUUCUGUUUAGGUGUCGUUAAAUGCCUUUUGCCAACAAGUCGUUGGCGACAACAGGAACGGCA ..((((.((-((((..((((..(.((((.(((((.((((........))))))))).)))).)..))))((((((.....)).....((((((.....)))))))))))))))))))).. ( -41.00) >DroSec_CAF1 9581 119 + 1 UCCUGUCUU-CUGUAUCCUGUCCCGGCGCUGUCCUGUUCUCCUGGCUGGGCGGACGUCUUCUGUUUAGGUGUCGUUAAAUGCCUUUUGCCAACAAGUCGUUGGCGACAACAGGAACGGCA .........-((((.((((((...((((((((((.(((.....))).)))))).))))...((((.((((((......))))))...((((((.....)))))))))))))))))))).. ( -40.40) >DroSim_CAF1 1501 119 + 1 UCCUGUCUU-CUGUAUCCUGUCCCGGUGCUGUCCUGUUCUCCUGGCUGGGCGGACGUCUUCUGUUUAGGUGUCGUUAAAUGCCUUUUGCCAACAAGUCGUUGGCGACAACAGGAACGGCA .........-................((((((((((((.((..(((..((((.((..((.......))..))))))....)))....((((((.....)))))))).)))))).)))))) ( -38.40) >DroEre_CAF1 9583 119 + 1 UCCUGUCUC-CAGUAUCCUGUCCCGCUGCUGUCCUGUUCUCCUGGCUGGGCGGACGUCUUCUGUUUAGGUGUCGUUAAAUGCCUUUUGCCAACAAGUCGUUGGCGACAACAGGAACGGCA ..(((....-)))..............(((((((((((.((..(((..((((.((..((.......))..))))))....)))....((((((.....)))))))).)))))).))))). ( -38.80) >DroYak_CAF1 6908 120 + 1 UCCUGUCUCCCAGUAUCCUGUCGCGAUGCUGUCCUGUUCUCCUGGCUGGGCGGACGUCUUCUGUUUAGGUGUCGUUAAAUGCCUUUUGCCAACAAGUCGUUGGCGACAACAGGAAGGGCA ....((((..((((((((....).)))))))(((((((.((..(((..((((.((..((.......))..))))))....)))....((((((.....)))))))).))))))).)))). ( -42.30) >consensus UCCUGUCUU_CUGUAUCCUGUCCCGGUGCUGUCCUGUUCUCCUGGCUGGGCGGACGUCUUCUGUUUAGGUGUCGUUAAAUGCCUUUUGCCAACAAGUCGUUGGCGACAACAGGAACGGCA ...........................(((((((((((.((..(((..((((.((..((.......))..))))))....)))....((((((.....)))))))).)))))).))))). (-36.28 = -36.68 + 0.40)


| Location | 7,196,041 – 7,196,148 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 92.93 |
| Mean single sequence MFE | -40.80 |
| Consensus MFE | -35.02 |
| Energy contribution | -35.42 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7196041 107 + 23771897 CCUAGCUGGGCGGACGUCUUCUGUUUAGGUGUCGUUAAAUGCCUUUUGCCAACAAGUCGUUGGCGACAACAGGAACGGCACACCAAAUGGCUCAGACACCAUGGGUG---------- .....(((((((((.....)))))))))((((((((...((....((((((((.....))))))))...))..))))))))(((..((((........)))).))).---------- ( -37.30) >DroSec_CAF1 9620 107 + 1 CCUGGCUGGGCGGACGUCUUCUGUUUAGGUGUCGUUAAAUGCCUUUUGCCAACAAGUCGUUGGCGACAACAGGAACGGCACACCAAAUGGCUCAGACACCAUGGCUG---------- ((((((((((((((.....)))))))))((((((((...((....((((((((.....))))))))...))..)))))))).))).((((........))))))...---------- ( -36.70) >DroSim_CAF1 1540 117 + 1 CCUGGCUGGGCGGACGUCUUCUGUUUAGGUGUCGUUAAAUGCCUUUUGCCAACAAGUCGUUGGCGACAACAGGAACGGCACACCAAAUGGCUCAGACACCAUGGGUGUGGCGACUAA ((((.(((((((((.....))))))))).((((((.....)).....((((((.....)))))))))).))))..((.((((((..((((........)))).)))))).))..... ( -44.80) >DroEre_CAF1 9622 117 + 1 CCUGGCUGGGCGGACGUCUUCUGUUUAGGUGUCGUUAAAUGCCUUUUGCCAACAAGUCGUUGGCGACAACAGGAACGGCACACCAAAUGGCUCAGACACCAUGGGUGUGGCAGCUAA ..((((((((((((.....)))))))))((((((((...((....((((((((.....))))))))...))..)))))))).)))..((((((..(((((...))))).).))))). ( -44.20) >DroYak_CAF1 6948 117 + 1 CCUGGCUGGGCGGACGUCUUCUGUUUAGGUGUCGUUAAAUGCCUUUUGCCAACAAGUCGUUGGCGACAACAGGAAGGGCACACCAAAUGGCUCAGACACCAUGGGUGUGGUGAUUGA .(((((((((((((.....))))))).(((((.(((.....(((.((((((((.....))))))))....)))...))))))))....))).))).((((((....))))))..... ( -41.00) >consensus CCUGGCUGGGCGGACGUCUUCUGUUUAGGUGUCGUUAAAUGCCUUUUGCCAACAAGUCGUUGGCGACAACAGGAACGGCACACCAAAUGGCUCAGACACCAUGGGUGUGG____U_A ((((((((((((((.....)))))))))((((((((...((....((((((((.....))))))))...))..)))))))).))).((((........))))))............. (-35.02 = -35.42 + 0.40)



| Location | 7,196,081 – 7,196,176 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 85.52 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -20.84 |
| Energy contribution | -22.16 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7196081 95 + 23771897 GCCUUUUGCCAACAAGUCGUUGGCGACAACAGGAACGGCACACCAAAUGGCUCAGACACCAUGGGUG---------------UGAUUAUUCAAAUAUUUAAACAAAAUCA .....((((((((.....))))))))......(((..(((((((..((((........)))).))))---------------)).)..)))................... ( -26.10) >DroSec_CAF1 9660 95 + 1 GCCUUUUGCCAACAAGUCGUUGGCGACAACAGGAACGGCACACCAAAUGGCUCAGACACCAUGGCUG---------------UGGUUAUUAAAAUAUUUAAACAAAAGCA .(((.((((((((.....))))))))....))).....(((((((..(((........)))))).))---------------)).......................... ( -21.30) >DroSim_CAF1 1580 110 + 1 GCCUUUUGCCAACAAGUCGUUGGCGACAACAGGAACGGCACACCAAAUGGCUCAGACACCAUGGGUGUGGCGACUAAAAGUGUGGUUAUUCAAAUAUUUAAACAAAAACA .(((.((((((((.....))))))))....)))..((.((((((..((((........)))).)))))).)).....((((((..........))))))........... ( -30.70) >DroEre_CAF1 9662 109 + 1 GCCUUUUGCCAACAAGUCGUUGGCGACAACAGGAACGGCACACCAAAUGGCUCAGACACCAUGGGUGUGGCAGCUAAAAGUUUUG-UAUCAAAAUAUUAAAACAAAAUCC .....((((((((.....)))))))).....(((..(.((((((..((((........)))).)))))).)........((((((-(((....))).))))))....))) ( -30.50) >DroYak_CAF1 6988 109 + 1 GCCUUUUGCCAACAAGUCGUUGGCGACAACAGGAAGGGCACACCAAAUGGCUCAGACACCAUGGGUGUGGUGAUUGAAAGUUUUG-UAUUCAAAUAUUAAAACAAAAGCU (((((((((((((.....))))))..(....)))))))).........(((((((.((((((....)))))).))))..((((((-(((....))).))))))....))) ( -31.60) >consensus GCCUUUUGCCAACAAGUCGUUGGCGACAACAGGAACGGCACACCAAAUGGCUCAGACACCAUGGGUGUGG____U_AAAGU_UGGUUAUUCAAAUAUUUAAACAAAAGCA .(((.((((((((.....))))))))....)))...(.((((((..((((........)))).)))))).)....................................... (-20.84 = -22.16 + 1.32)



| Location | 7,196,081 – 7,196,176 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 85.52 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -19.92 |
| Energy contribution | -21.24 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7196081 95 - 23771897 UGAUUUUGUUUAAAUAUUUGAAUAAUCA---------------CACCCAUGGUGUCUGAGCCAUUUGGUGUGCCGUUCCUGUUGUCGCCAACGACUUGUUGGCAAAAGGC .(((.((((((((....)))))))).((---------------((((.(((((......)))))..))))))...........)))((((((.....))))))....... ( -26.30) >DroSec_CAF1 9660 95 - 1 UGCUUUUGUUUAAAUAUUUUAAUAACCA---------------CAGCCAUGGUGUCUGAGCCAUUUGGUGUGCCGUUCCUGUUGUCGCCAACGACUUGUUGGCAAAAGGC .((((((.................((.(---------------(((..((((..(....(((....))))..))))..)))).)).((((((.....)))))).)))))) ( -25.70) >DroSim_CAF1 1580 110 - 1 UGUUUUUGUUUAAAUAUUUGAAUAACCACACUUUUAGUCGCCACACCCAUGGUGUCUGAGCCAUUUGGUGUGCCGUUCCUGUUGUCGCCAACGACUUGUUGGCAAAAGGC (((..((((((((....))))))))..)))(((((...((.((((((.(((((......)))))..)))))).))...........((((((.....))))))))))).. ( -31.80) >DroEre_CAF1 9662 109 - 1 GGAUUUUGUUUUAAUAUUUUGAUA-CAAAACUUUUAGCUGCCACACCCAUGGUGUCUGAGCCAUUUGGUGUGCCGUUCCUGUUGUCGCCAACGACUUGUUGGCAAAAGGC ...((((((.((((....)))).)-)))))((((((((.((.(((((.(((((......)))))..))))))).))).........((((((.....))))))))))).. ( -31.80) >DroYak_CAF1 6988 109 - 1 AGCUUUUGUUUUAAUAUUUGAAUA-CAAAACUUUCAAUCACCACACCCAUGGUGUCUGAGCCAUUUGGUGUGCCCUUCCUGUUGUCGCCAACGACUUGUUGGCAAAAGGC .((((((....(((((.(((((..-.......)))))....((((((.(((((......)))))..)))))).......)))))..((((((.....)))))).)))))) ( -28.90) >consensus UGAUUUUGUUUAAAUAUUUGAAUAACCA_ACUUU_A____CCACACCCAUGGUGUCUGAGCCAUUUGGUGUGCCGUUCCUGUUGUCGCCAACGACUUGUUGGCAAAAGGC .((((((..................................((((((.(((((......)))))..))))))..............((((((.....)))))).)))))) (-19.92 = -21.24 + 1.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:30 2006