
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,195,137 – 7,195,309 |
| Length | 172 |
| Max. P | 0.999936 |

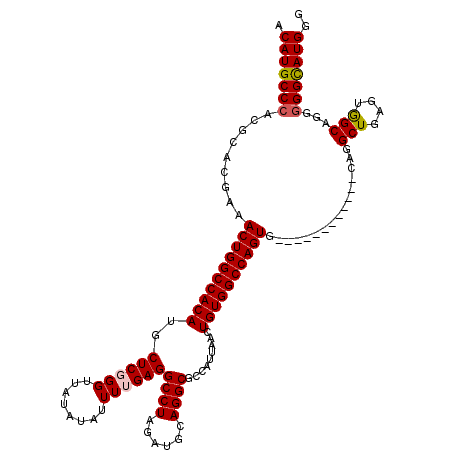
| Location | 7,195,137 – 7,195,245 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.69 |
| Mean single sequence MFE | -46.28 |
| Consensus MFE | -42.14 |
| Energy contribution | -42.42 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.66 |
| SVM RNA-class probability | 0.999936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
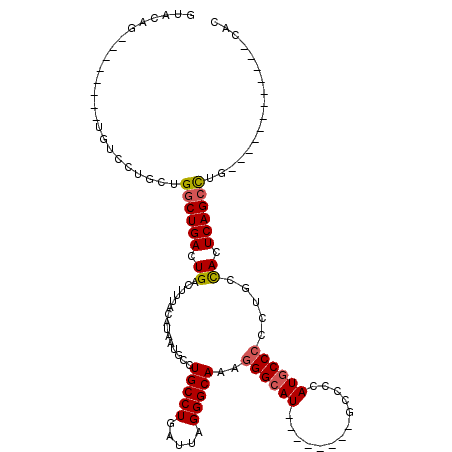
>3L_DroMel_CAF1 7195137 108 + 23771897 ACAUGCCCAUACACGUAACUGGCCACAUGCUCGGGUUAUAUAUUUUGAGGCCUAGAUGCAGGCGCCAUUAACUGUGGCCAGUA------------CAGGCUGACUGGCAGGGGGUAUGGG .(((((((......(((.(((((((((..((((((........))))))((((......)))).........)))))))))))------------)..(((....)))...))))))).. ( -43.90) >DroSec_CAF1 8706 108 + 1 ACAUGCCCACGCACGAAACUGGCCACAUGCUCGGGUUAUAUAUUUUGAGGCCUAGAUGCAGGCGCCAUUAACUGUGGCCAGUG------------CAGGCUGAGUGGCAGGGGGCAUGGG .(((((((.....(...((((((((((..((((((........))))))((((......)))).........)))))))))).------------..)(((....)))...))))))).. ( -45.50) >DroSim_CAF1 629 107 + 1 ACAUGCCCACGCACGAAACUGGCCACAUGCUCGGGUUAUAUAUUUUGAGGCCUAGAUGCAGGCGCCAUUAACUGUGGCCAGUG------------CAGGCUGAGUGGCAGG-GGCAUGGG .(((((((.....(...((((((((((..((((((........))))))((((......)))).........)))))))))).------------..)(((....)))..)-)))))).. ( -45.50) >DroEre_CAF1 8780 107 + 1 ACAUGCCCACGCACGAAACUGGCCACAUGCUCUGGUUAUAUAUUUUGAGGCCUAGAUGCAGGCGCCAUUAACUGUGGCCAGUG------------CAAGCUGAGUGGCA-GGGGCAUGGC .(((((((..((((.....(((((((((((((((((..((.....))..))).))).)))(....)......)))))))))))------------)..(((....))).-.))))))).. ( -43.70) >DroYak_CAF1 6023 120 + 1 ACAUGCCCACGCACGAAACUGGCCACAUGCUCUGGUUAUAUAUUUUGAGGCCUAGAUGCAGGCGCCAUUAACUGUGGCCAGUGCAGUGCCAGUUGCAAGCUGAGUAGCAUGGGGCAUGGC .(((((((..((((...(((((((((((((((((((..((.....))..))).))).)))(....)......))))))))))...))))(((((((.......))))).))))))))).. ( -52.80) >consensus ACAUGCCCACGCACGAAACUGGCCACAUGCUCGGGUUAUAUAUUUUGAGGCCUAGAUGCAGGCGCCAUUAACUGUGGCCAGUG____________CAGGCUGAGUGGCAGGGGGCAUGGG .(((((((.........((((((((((..((((((........))))))((((......)))).........))))))))))................(((....)))...))))))).. (-42.14 = -42.42 + 0.28)


| Location | 7,195,137 – 7,195,245 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.69 |
| Mean single sequence MFE | -40.80 |
| Consensus MFE | -34.75 |
| Energy contribution | -35.15 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.999071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7195137 108 - 23771897 CCCAUACCCCCUGCCAGUCAGCCUG------------UACUGGCCACAGUUAAUGGCGCCUGCAUCUAGGCCUCAAAAUAUAUAACCCGAGCAUGUGGCCAGUUACGUGUAUGGGCAUGU (((((((...(((.....)))...(------------((((((((((((((..(((.(((((....))))).)))..............))).))))))))).)))..)))))))..... ( -35.89) >DroSec_CAF1 8706 108 - 1 CCCAUGCCCCCUGCCACUCAGCCUG------------CACUGGCCACAGUUAAUGGCGCCUGCAUCUAGGCCUCAAAAUAUAUAACCCGAGCAUGUGGCCAGUUUCGUGCGUGGGCAUGU ..(((((((...(((((...(....------------)(((((((((((((..(((.(((((....))))).)))..............))).))))))))))...))).))))))))). ( -39.09) >DroSim_CAF1 629 107 - 1 CCCAUGCC-CCUGCCACUCAGCCUG------------CACUGGCCACAGUUAAUGGCGCCUGCAUCUAGGCCUCAAAAUAUAUAACCCGAGCAUGUGGCCAGUUUCGUGCGUGGGCAUGU ..((((((-(.(((.((...(....------------)(((((((((((((..(((.(((((....))))).)))..............))).))))))))))...))))).))))))). ( -40.69) >DroEre_CAF1 8780 107 - 1 GCCAUGCCCC-UGCCACUCAGCUUG------------CACUGGCCACAGUUAAUGGCGCCUGCAUCUAGGCCUCAAAAUAUAUAACCAGAGCAUGUGGCCAGUUUCGUGCGUGGGCAUGU ..(((((((.-(((.((...(....------------)((((((((((.....(((.(((((....)))))..............))).....))))))))))...))))).))))))). ( -40.86) >DroYak_CAF1 6023 120 - 1 GCCAUGCCCCAUGCUACUCAGCUUGCAACUGGCACUGCACUGGCCACAGUUAAUGGCGCCUGCAUCUAGGCCUCAAAAUAUAUAACCAGAGCAUGUGGCCAGUUUCGUGCGUGGGCAUGU ..(((((((.((((.((...((.(((.....)))..))((((((((((.....(((.(((((....)))))..............))).....))))))))))...))))))))))))). ( -47.46) >consensus CCCAUGCCCCCUGCCACUCAGCCUG____________CACUGGCCACAGUUAAUGGCGCCUGCAUCUAGGCCUCAAAAUAUAUAACCCGAGCAUGUGGCCAGUUUCGUGCGUGGGCAUGU ..(((((((...((......))................(((((((((((((..(((.(((((....))))).)))..............))).)))))))))).........))))))). (-34.75 = -35.15 + 0.40)



| Location | 7,195,177 – 7,195,277 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.87 |
| Mean single sequence MFE | -45.08 |
| Consensus MFE | -36.88 |
| Energy contribution | -37.52 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.82 |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.998476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7195177 100 + 23771897 UAUUUUGAGGCCUAGAUGCAGGCGCCAUUAACUGUGGCCAGUA------------CAGGCUGACUGGCAGGGGGUAUGGGGC--------AUGCCCUUUGCCCUAAUCAGGCAGGCAUUA .........((((......))))(((....((((....)))).------------..(.((((..(((((((((((((...)--------))))))))))))....)))).).))).... ( -40.20) >DroSec_CAF1 8746 108 + 1 UAUUUUGAGGCCUAGAUGCAGGCGCCAUUAACUGUGGCCAGUG------------CAGGCUGAGUGGCAGGGGGCAUGGGGCAUGAUGGCAUGCCCUUUGCCCUAAUCAGGCAGGCAUUA .........((((...((((.(.(((((.....))))))..))------------))(.((((.......((((((.((((((((....)))))))).))))))..)))).))))).... ( -50.00) >DroSim_CAF1 669 107 + 1 UAUUUUGAGGCCUAGAUGCAGGCGCCAUUAACUGUGGCCAGUG------------CAGGCUGAGUGGCAGG-GGCAUGGGGCAUGAUGGCAUGCCCUUUGCCCUAAUCAGGCAGGCAUUA .........((((...((((.(.(((((.....))))))..))------------))(.((((......((-((((.((((((((....)))))))).))))))..)))).))))).... ( -49.50) >DroEre_CAF1 8820 99 + 1 UAUUUUGAGGCCUAGAUGCAGGCGCCAUUAACUGUGGCCAGUG------------CAAGCUGAGUGGCA-GGGGCAUGGCUC--------AUGCCCUUUGCCCUAAUCAGGCAGGCAUUA .........((((...((((.(.(((((.....))))))..))------------)).(((.((.((((-((((((((...)--------)))))).))))))).....))))))).... ( -41.00) >DroYak_CAF1 6063 112 + 1 UAUUUUGAGGCCUAGAUGCAGGCGCCAUUAACUGUGGCCAGUGCAGUGCCAGUUGCAAGCUGAGUAGCAUGGGGCAUGGCUG--------AUGCCCUUUGCCCUAAUCAGGCAGGCAUUA .........((((......))))(((((.....))))).(((((..((((.(((((.......))))).(((((((.(((..--------..)))...)))))))....)))).))))). ( -44.70) >consensus UAUUUUGAGGCCUAGAUGCAGGCGCCAUUAACUGUGGCCAGUG____________CAGGCUGAGUGGCAGGGGGCAUGGGGC________AUGCCCUUUGCCCUAAUCAGGCAGGCAUUA ..............(((((..(((((((.....))))).....................((((..((((((((((((.............))))))))))))....))))))..))))). (-36.88 = -37.52 + 0.64)

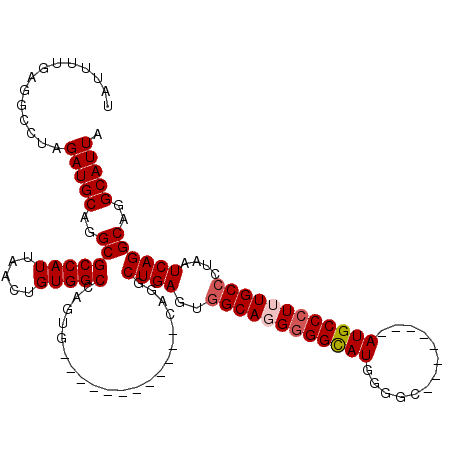

| Location | 7,195,177 – 7,195,277 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.87 |
| Mean single sequence MFE | -35.74 |
| Consensus MFE | -29.14 |
| Energy contribution | -29.38 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7195177 100 - 23771897 UAAUGCCUGCCUGAUUAGGGCAAAGGGCAU--------GCCCCAUACCCCCUGCCAGUCAGCCUG------------UACUGGCCACAGUUAAUGGCGCCUGCAUCUAGGCCUCAAAAUA ..((((((((((.....))))...))))))--------(((....((.....(((((((.....)------------.))))))....))....)))(((((....)))))......... ( -31.60) >DroSec_CAF1 8746 108 - 1 UAAUGCCUGCCUGAUUAGGGCAAAGGGCAUGCCAUCAUGCCCCAUGCCCCCUGCCACUCAGCCUG------------CACUGGCCACAGUUAAUGGCGCCUGCAUCUAGGCCUCAAAAUA ........(((((....(((((..(((((((....)))))))..)))))..(((......(((..------------.((((....))))....)))....)))..)))))......... ( -38.30) >DroSim_CAF1 669 107 - 1 UAAUGCCUGCCUGAUUAGGGCAAAGGGCAUGCCAUCAUGCCCCAUGCC-CCUGCCACUCAGCCUG------------CACUGGCCACAGUUAAUGGCGCCUGCAUCUAGGCCUCAAAAUA ........(((((....(((((..(((((((....)))))))..))))-).(((......(((..------------.((((....))))....)))....)))..)))))......... ( -38.30) >DroEre_CAF1 8820 99 - 1 UAAUGCCUGCCUGAUUAGGGCAAAGGGCAU--------GAGCCAUGCCCC-UGCCACUCAGCUUG------------CACUGGCCACAGUUAAUGGCGCCUGCAUCUAGGCCUCAAAAUA ....(((.((((((....((((..((((((--------(...))))))).-))))..))))...)------------)((((....))))....)))(((((....)))))......... ( -34.80) >DroYak_CAF1 6063 112 - 1 UAAUGCCUGCCUGAUUAGGGCAAAGGGCAU--------CAGCCAUGCCCCAUGCUACUCAGCUUGCAACUGGCACUGCACUGGCCACAGUUAAUGGCGCCUGCAUCUAGGCCUCAAAAUA ....(((((.((((....((((..((((((--------.....))))))..))))..))))..((((...(((.(((.((((....))))...))).)))))))..)))))......... ( -35.70) >consensus UAAUGCCUGCCUGAUUAGGGCAAAGGGCAU________GCCCCAUGCCCCCUGCCACUCAGCCUG____________CACUGGCCACAGUUAAUGGCGCCUGCAUCUAGGCCUCAAAAUA ....(((...((((....((((..((((((.............))))))..))))..)))).................((((....))))....)))(((((....)))))......... (-29.14 = -29.38 + 0.24)

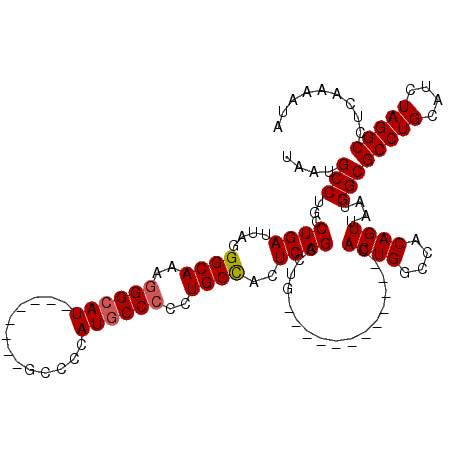

| Location | 7,195,217 – 7,195,309 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.86 |
| Mean single sequence MFE | -44.44 |
| Consensus MFE | -28.82 |
| Energy contribution | -30.66 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.97 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7195217 92 + 23771897 GUA------------CAGGCUGACUGGCAGGGGGUAUGGGGC--------AUGCCCUUUGCCCUAAUCAGGCAGGCAUUAUGUAAAGUCAGUCAGCCAGCAGGACA--------CUGUAC ...------------..(((((((((((..((((((.((((.--------...)))).))))))......(((.......)))...))))))))))).((((....--------)))).. ( -43.90) >DroSec_CAF1 8786 100 + 1 GUG------------CAGGCUGAGUGGCAGGGGGCAUGGGGCAUGAUGGCAUGCCCUUUGCCCUAAUCAGGCAGGCAUUAUGUAAAGUCAGUCAGCCAGCAGGACA--------CUGUAC ...------------..((((((.((((..((((((.((((((((....)))))))).))))))......(((.......)))...)))).)))))).((((....--------)))).. ( -49.60) >DroSim_CAF1 709 99 + 1 GUG------------CAGGCUGAGUGGCAGG-GGCAUGGGGCAUGAUGGCAUGCCCUUUGCCCUAAUCAGGCAGGCAUUAUGUAAAGUCAGUCAGCCAGCAGGACA--------CUGUAC ...------------..((((((.((((.((-((((.((((((((....)))))))).))))))......(((.......)))...)))).)))))).((((....--------)))).. ( -49.50) >DroEre_CAF1 8860 91 + 1 GUG------------CAAGCUGAGUGGCA-GGGGCAUGGCUC--------AUGCCCUUUGCCCUAAUCAGGCAGGCAUUAUGUAAAGUCAGUCAGGCAGCAGGACA--------CUGUAC .((------------(...((((.((((.-((((((((...)--------)))))))(((((.......)))))............)))).)))))))((((....--------)))).. ( -33.80) >DroYak_CAF1 6103 112 + 1 GUGCAGUGCCAGUUGCAAGCUGAGUAGCAUGGGGCAUGGCUG--------AUGCCCUUUGCCCUAAUCAGGCAGGCAUUAUGUAAAGUCAGUCAGCCAGCAGGACACUGCCACACUGUAC ((((((((((((((((.......))))).)))(((.((((((--------(((((...((((.......))))))))..........)))))))))).((((....))))..)))))))) ( -45.40) >consensus GUG____________CAGGCUGAGUGGCAGGGGGCAUGGGGC________AUGCCCUUUGCCCUAAUCAGGCAGGCAUUAUGUAAAGUCAGUCAGCCAGCAGGACA________CUGUAC .................((((((.((((..((((((.(((((..........))))).))))))......(((.......)))...)))).)))))).((((............)))).. (-28.82 = -30.66 + 1.84)


| Location | 7,195,217 – 7,195,309 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.86 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -21.28 |
| Energy contribution | -21.48 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7195217 92 - 23771897 GUACAG--------UGUCCUGCUGGCUGACUGACUUUACAUAAUGCCUGCCUGAUUAGGGCAAAGGGCAU--------GCCCCAUACCCCCUGCCAGUCAGCCUG------------UAC ((((((--------(.....)))(((((((((..........((((((((((.....))))...))))))--------((............))))))))))).)------------))) ( -31.80) >DroSec_CAF1 8786 100 - 1 GUACAG--------UGUCCUGCUGGCUGACUGACUUUACAUAAUGCCUGCCUGAUUAGGGCAAAGGGCAUGCCAUCAUGCCCCAUGCCCCCUGCCACUCAGCCUG------------CAC ......--------.....(((.((((((.((........((((.........))))(((((..(((((((....)))))))..))))).....)).)))))).)------------)). ( -35.30) >DroSim_CAF1 709 99 - 1 GUACAG--------UGUCCUGCUGGCUGACUGACUUUACAUAAUGCCUGCCUGAUUAGGGCAAAGGGCAUGCCAUCAUGCCCCAUGCC-CCUGCCACUCAGCCUG------------CAC ......--------.....(((.((((((.((........((((.........))))(((((..(((((((....)))))))..))))-)....)).)))))).)------------)). ( -35.30) >DroEre_CAF1 8860 91 - 1 GUACAG--------UGUCCUGCUGCCUGACUGACUUUACAUAAUGCCUGCCUGAUUAGGGCAAAGGGCAU--------GAGCCAUGCCCC-UGCCACUCAGCUUG------------CAC (((.((--------((((.........)))..))).)))....(((..((.(((....((((..((((((--------(...))))))).-))))..)))))..)------------)). ( -27.70) >DroYak_CAF1 6103 112 - 1 GUACAGUGUGGCAGUGUCCUGCUGGCUGACUGACUUUACAUAAUGCCUGCCUGAUUAGGGCAAAGGGCAU--------CAGCCAUGCCCCAUGCUACUCAGCUUGCAACUGGCACUGCAC ..........((((((((.(((.((((((................((((......))))(((..((((((--------.....))))))..)))...)))))).)))...)))))))).. ( -41.40) >consensus GUACAG________UGUCCUGCUGGCUGACUGACUUUACAUAAUGCCUGCCUGAUUAGGGCAAAGGGCAU________GCCCCAUGCCCCCUGCCACUCAGCCUG____________CAC .......................((((((.((...............(((((.....)))))..((((((.............)))))).....)).))))))................. (-21.28 = -21.48 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:24 2006