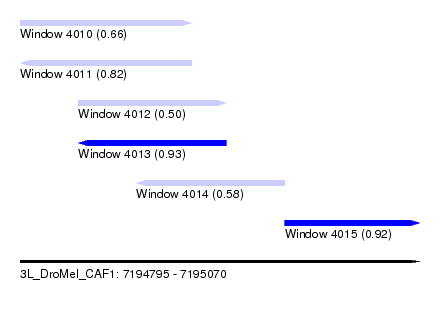
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,194,795 – 7,195,070 |
| Length | 275 |
| Max. P | 0.931874 |

| Location | 7,194,795 – 7,194,913 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.84 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -24.10 |
| Energy contribution | -24.23 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7194795 118 + 23771897 AGGGAAAAGGAGAAAGAAUAGAAAAAACGUCAGCAGGCCAUACAAAACAAAUACCAUUUCCGUUGCCUACUUCGAGGCGUGCGUAUACGUAAUAUCUG-UGUGCGUAU-ACGUAUACGCA ..(((((.((..........(......)(((....)))...............)).)))))...((((......)))).((((((((((((.(((...-.....))))-))))))))))) ( -28.80) >DroSec_CAF1 8376 120 + 1 AGGAAAAAGGAGAAAGAAUAGAAAAAACGUCAGCAGACCAUACAAAACAAAUUCCAUUUCCGUUGCCUACUUCGAGGCGUGCGUAUACGUAAUAUCUGCUGUGCGUAUAACGUAUACGCA .(((((..((((........(......)(((....))).............)))).)))))...((((......)))).(((((((((((.((((..((...)))))).))))))))))) ( -30.20) >DroSim_CAF1 284 119 + 1 AGGGAAAAGAAGAAAGAAUUGAAAAAACGUCAGCAGACCAUACAAAACAAAUACCAUUUCCGUUGCCUAUUUCGAGGCGUGCGUAUACGUAAUAUCUGCUGUGCGUAU-ACGUAUACGCA ..(((((...........((....))..(((....)))..................)))))...((((......)))).((((((((((((.(((..((...))))))-))))))))))) ( -27.30) >DroYak_CAF1 5647 118 + 1 AUGGAAAAGAAGAAAGAAUAGUAAAAACGUCAGCAAACCAUACAAAACAAAUACCAUUUCCGUUGCCUACUUCGAGGCGUGCGUAUACGUAAUAUCU-CUGUGCGUAU-ACGUAUACGCA (((((((.............((....))(....)......................))))))).((((......)))).((((((((((((.(((..-......))))-))))))))))) ( -25.30) >consensus AGGGAAAAGAAGAAAGAAUAGAAAAAACGUCAGCAGACCAUACAAAACAAAUACCAUUUCCGUUGCCUACUUCGAGGCGUGCGUAUACGUAAUAUCUGCUGUGCGUAU_ACGUAUACGCA .(((((..............(......)(((....)))..................)))))...((((......)))).(((((((((((.(((((......).)))).))))))))))) (-24.10 = -24.23 + 0.12)


| Location | 7,194,795 – 7,194,913 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.84 |
| Mean single sequence MFE | -30.43 |
| Consensus MFE | -26.96 |
| Energy contribution | -26.53 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7194795 118 - 23771897 UGCGUAUACGU-AUACGCACA-CAGAUAUUACGUAUACGCACGCCUCGAAGUAGGCAACGGAAAUGGUAUUUGUUUUGUAUGGCCUGCUGACGUUUUUUCUAUUCUUUCUCCUUUUCCCU (((((((((((-(....(...-..)....)))))))))))).....((.(((((((.((((((..........))))))...)))))))..))........................... ( -30.10) >DroSec_CAF1 8376 120 - 1 UGCGUAUACGUUAUACGCACAGCAGAUAUUACGUAUACGCACGCCUCGAAGUAGGCAACGGAAAUGGAAUUUGUUUUGUAUGGUCUGCUGACGUUUUUUCUAUUCUUUCUCCUUUUUCCU (((((((((((.((((........).))).))))))))))).((((......))))...(((((.(((..............(((....)))(......).........)))..))))). ( -30.40) >DroSim_CAF1 284 119 - 1 UGCGUAUACGU-AUACGCACAGCAGAUAUUACGUAUACGCACGCCUCGAAAUAGGCAACGGAAAUGGUAUUUGUUUUGUAUGGUCUGCUGACGUUUUUUCAAUUCUUUCUUCUUUUCCCU (((((((((((-(....(......)....)))))))))))).((((......))))...(((((.((.....(..(((....(((....))).......)))..).....)).))))).. ( -30.00) >DroYak_CAF1 5647 118 - 1 UGCGUAUACGU-AUACGCACAG-AGAUAUUACGUAUACGCACGCCUCGAAGUAGGCAACGGAAAUGGUAUUUGUUUUGUAUGGUUUGCUGACGUUUUUACUAUUCUUUCUUCUUUUCCAU (((((((((((-(....(....-.)....)))))))))))).((((......))))...((((((((((..((((..(((.....))).))))....))))))...........)))).. ( -31.20) >consensus UGCGUAUACGU_AUACGCACAGCAGAUAUUACGUAUACGCACGCCUCGAAGUAGGCAACGGAAAUGGUAUUUGUUUUGUAUGGUCUGCUGACGUUUUUUCUAUUCUUUCUCCUUUUCCCU (((((((((((.((((........).))).))))))))))).((((......))))...(((((.((.....(((..(((.....))).)))(......)..........)).))))).. (-26.96 = -26.53 + -0.44)

| Location | 7,194,835 – 7,194,937 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -26.24 |
| Consensus MFE | -24.80 |
| Energy contribution | -24.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.95 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7194835 102 + 23771897 UACAAAACAAAUACCAUUUCCGUUGCCUACUUCGAGGCGUGCGUAUACGUAAUAUCUG-UGUGCGUAU-ACGUAUACGCAAUAUCUGAAUGCAUGGCUGUGUGC ......(((....((((...((((((((......)))).((((((((((((.(((...-.....))))-))))))))))).......)))).))))...))).. ( -27.90) >DroSec_CAF1 8416 102 + 1 UACAAAACAAAUUCCAUUUCCGUUGCCUACUUCGAGGCGUGCGUAUACGUAAUAUCUGCUGUGCGUAUAACGUAUACGCAAUAUCUGAAUGCAUGCCUGUGU-- ((((......((((..........((((......)))).(((((((((((.((((..((...)))))).)))))))))))......)))).......)))).-- ( -25.72) >DroSim_CAF1 324 103 + 1 UACAAAACAAAUACCAUUUCCGUUGCCUAUUUCGAGGCGUGCGUAUACGUAAUAUCUGCUGUGCGUAU-ACGUAUACGCAAUAUCUGAAUGCAUGCCUGUGUGC .......((.(((...........((((......)))).((((((((((((.(((..((...))))))-))))))))))).))).))...(((((....))))) ( -25.10) >consensus UACAAAACAAAUACCAUUUCCGUUGCCUACUUCGAGGCGUGCGUAUACGUAAUAUCUGCUGUGCGUAU_ACGUAUACGCAAUAUCUGAAUGCAUGCCUGUGUGC ......(((.....(((((.....((((......)))).(((((((((((.(((((......).)))).)))))))))))......)))))......))).... (-24.80 = -24.80 + 0.00)

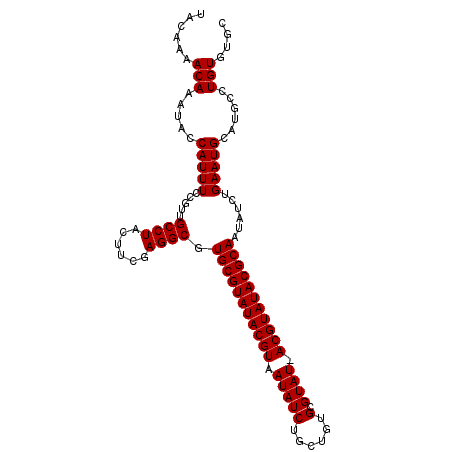

| Location | 7,194,835 – 7,194,937 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -27.93 |
| Energy contribution | -28.27 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7194835 102 - 23771897 GCACACAGCCAUGCAUUCAGAUAUUGCGUAUACGU-AUACGCACA-CAGAUAUUACGUAUACGCACGCCUCGAAGUAGGCAACGGAAAUGGUAUUUGUUUUGUA (((.((((((((...(((......(((((((((((-(....(...-..)....)))))))))))).((((......))))...))).)))))...)))..))). ( -29.60) >DroSec_CAF1 8416 102 - 1 --ACACAGGCAUGCAUUCAGAUAUUGCGUAUACGUUAUACGCACAGCAGAUAUUACGUAUACGCACGCCUCGAAGUAGGCAACGGAAAUGGAAUUUGUUUUGUA --(((.(((((...((((......(((((((((((.((((........).))).))))))))))).((((......))))..........)))).)))))))). ( -27.20) >DroSim_CAF1 324 103 - 1 GCACACAGGCAUGCAUUCAGAUAUUGCGUAUACGU-AUACGCACAGCAGAUAUUACGUAUACGCACGCCUCGAAAUAGGCAACGGAAAUGGUAUUUGUUUUGUA ((......)).((((..((((((((((((((((((-(....(......)....)))))))))))..((((......)))).........))))))))...)))) ( -30.80) >consensus GCACACAGGCAUGCAUUCAGAUAUUGCGUAUACGU_AUACGCACAGCAGAUAUUACGUAUACGCACGCCUCGAAGUAGGCAACGGAAAUGGUAUUUGUUUUGUA ...........((((..((((((((((((((((((.((((........).))).))))))))))..((((......)))).........))))))))...)))) (-27.93 = -28.27 + 0.33)


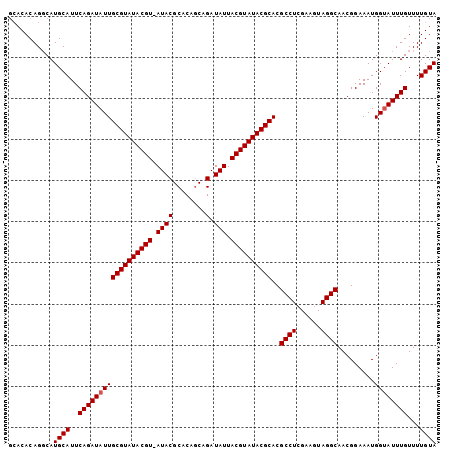
| Location | 7,194,875 – 7,194,977 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 92.28 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -21.80 |
| Energy contribution | -22.13 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7194875 102 - 23771897 CUGCAGCCGAUUAACUCUGGACCCUGCCAUACGCACACACGCACACAGCCAUGCAUUCAGAUAUUGCGUAUACGU-AUACGCACA-CAGAUAUUACGUAUACGC ...............((((((...(((.....))).....(((........))).))))))....((((((((((-(....(...-..)....))))))))))) ( -22.70) >DroSec_CAF1 8456 96 - 1 CUGCAGCCGAUUAACUCUGGACACUGCCAUACGC--------ACACAGGCAUGCAUUCAGAUAUUGCGUAUACGUUAUACGCACAGCAGAUAUUACGUAUACGC .(((((((.........(((......))).....--------.....))).))))..........((((((((((.((((........).))).)))))))))) ( -24.91) >DroSim_CAF1 364 103 - 1 CUGCAGCCGAUUAACUCUGGACCCUGCCAUACGCACACACGCACACAGGCAUGCAUUCAGAUAUUGCGUAUACGU-AUACGCACAGCAGAUAUUACGUAUACGC .(((((((.........(((......)))...((......)).....))).))))..........((((((((((-(....(......)....))))))))))) ( -27.80) >consensus CUGCAGCCGAUUAACUCUGGACCCUGCCAUACGCACACACGCACACAGGCAUGCAUUCAGAUAUUGCGUAUACGU_AUACGCACAGCAGAUAUUACGUAUACGC .(((((((((.....))(((......)))..................))).))))..........((((((((((.((((........).))).)))))))))) (-21.80 = -22.13 + 0.33)


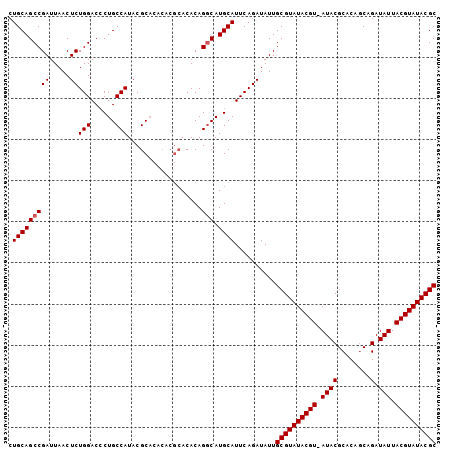
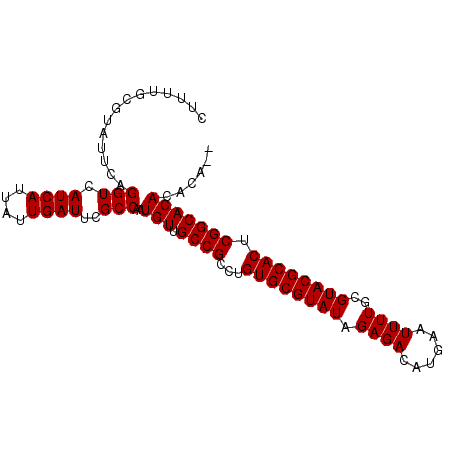
| Location | 7,194,977 – 7,195,070 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 96.47 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7194977 93 + 23771897 CUUUUGCGUAUUCAGGUCAUCAUUAUUGAUUCGCCAAUGUUGCCGCCUGUGCGUAUAGAGACAUGAAUUUUGCGUACGCACUCGGCACACACA-- ..............(((.((((....))))..)))..((((((((...((((((((.((((......))))..)))))))).))))).)))..-- ( -27.10) >DroSec_CAF1 8552 90 + 1 CUUUUGCGUAUUCAGGUCAUCAUUAUUGAUUCGCCAAUGUUGCCGCCUGUGCGUAUAGAGACAUGAAUUUUGCGUACGCACUCGGCACAC----- ..............(((.((((....))))..)))..(((.((((...((((((((.((((......))))..)))))))).))))))).----- ( -26.20) >DroSim_CAF1 467 95 + 1 CUUUUGCGUAUUCAGGUCAUCAUUAUUGAUUCGCCAAUGUUGCCGCCUGUGCGUAUAGAGACAUGAAUUUUGCGUACGCACUCGGCACACACACA .......((.....(((.((((....))))..)))..((((((((...((((((((.((((......))))..)))))))).))))).))).)). ( -27.20) >consensus CUUUUGCGUAUUCAGGUCAUCAUUAUUGAUUCGCCAAUGUUGCCGCCUGUGCGUAUAGAGACAUGAAUUUUGCGUACGCACUCGGCACACACA__ ..............(((.((((....))))..)))..(((.((((...((((((((.((((......))))..)))))))).)))))))...... (-26.20 = -26.20 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:18 2006