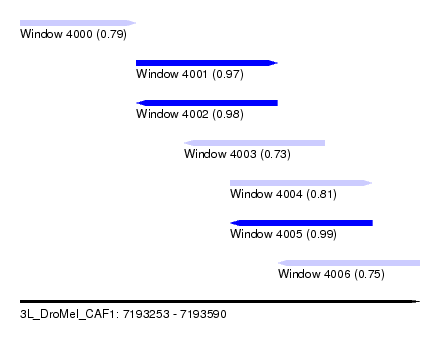
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,193,253 – 7,193,590 |
| Length | 337 |
| Max. P | 0.985436 |

| Location | 7,193,253 – 7,193,351 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 77.00 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -17.14 |
| Energy contribution | -18.25 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7193253 98 + 23771897 ACACACACAGAGCAAACACACAGGGCUGCAGCAAACGUAUAUGU----ACACACUUGUAUGCCCCGCAAGUCUGUGUGUGUGUGUGUGUGAUUGUUUGUCCG .(((((((...(((.((((((((((((((((.....(((....)----))....))))).))))(....)..))))))).))))))))))............ ( -33.30) >DroEre_CAF1 6667 86 + 1 ACACAUACAGUGCCAACACACAGGGCUGCAGCAAACGUAUAUGU--------------AUACCCAGUAAGUGUG--UAUGUGUGUGUGUGAUUGCUUGCCAG .........(((......)))..(((.((((((.((((((((((--------------((((.(.....).)))--))))))))))).)).))))..))).. ( -23.80) >DroYak_CAF1 3750 98 + 1 ACACAUACACAGCCAGCACACAGGGCUGCAGCAAAUGUAUAUGUAUGUAAACACUCGUAUACCCAGUAUGAGUG----UGUGUGUGUGUGAUUGUUUGCCCG .((((((((((((((((.......))))..))..................((((((((((.....)))))))))----).))))))))))............ ( -32.40) >consensus ACACAUACAGAGCCAACACACAGGGCUGCAGCAAACGUAUAUGU____A_ACACU_GUAUACCCAGUAAGUGUG__U_UGUGUGUGUGUGAUUGUUUGCCCG (((((((((.......(((((...((....))....(((((((............))))))).......)))))......)))))))))............. (-17.14 = -18.25 + 1.12)

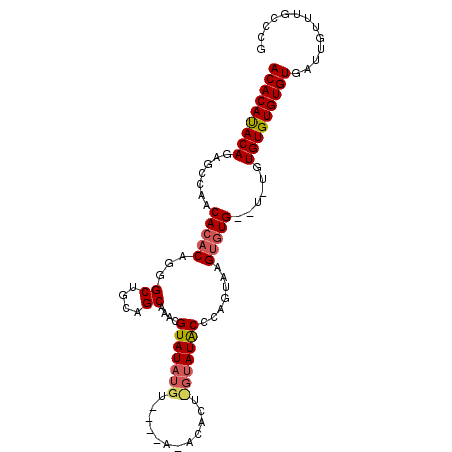
| Location | 7,193,351 – 7,193,470 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.72 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -27.06 |
| Energy contribution | -26.88 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7193351 119 + 23771897 UAGAAUGCAAUAAGGGGUCAGGCUUUAAGAGCCAUAACAAAUGGGUUAAGUAAGACCG-AAGAUGGAAAAUGGGCAUCCAAGGGAAGGGCAAAAAGUCGCCCUGUUAUGCAUAACAAAGA ....(((((......((((.(((((...))))).((((......)))).....)))).-............((((.(((...)))..(((.....))))))).....)))))........ ( -29.70) >DroSec_CAF1 6847 119 + 1 UAGAAUGCAAUAAGGGGUCAGGCUUUAAGAGCCAUAACAAAUGGGUUAAGUAAGACCG-AAGAUGGCAAAUGGACAUCCAAGGGAAGGGCAAAAAGUCGCCCCGUUAUGCAUAACAAAGA ....(((((....(((((..((((((....(((((......(.((((......)))).-)..)))))...(((....)))............)))))))))))....)))))........ ( -32.80) >DroEre_CAF1 6753 120 + 1 UAGAAUGCAAUAAGGGGUCAGGCUUUAAGAGCCAUAACAAAUGGGUUAAGUAAGACCGGAAGAUGGGGAAUGUGCAUCCAGGGUACGGGCAAAAAGUCGCCCUGUUAUGCAUAACAAAGA ....(((((......((((.(((((...))))).((((......)))).....))))....(((((((..(((((.......)))))(((.....))).))))))).)))))........ ( -32.00) >DroYak_CAF1 3848 119 + 1 UAGAAUGCAAUAAGGGGUCAGGCUUUAAGAGCCAUAACAAAUGGGUUAAGUAAGACCG-AAGAUGGGGAAUGUGCAUCCAGGGUAAGGGCAAAAAAUGGUCCUGCUAUGCAUAACAAAGA ....(((((......((((.(((((...))))).((((......)))).....)))).-..((((.(.....).))))...((((.((((........)))))))).)))))........ ( -28.40) >consensus UAGAAUGCAAUAAGGGGUCAGGCUUUAAGAGCCAUAACAAAUGGGUUAAGUAAGACCG_AAGAUGGGAAAUGGGCAUCCAAGGGAAGGGCAAAAAGUCGCCCUGUUAUGCAUAACAAAGA ....(((((......((((.(((((...))))).((((......)))).....))))....((((.........))))........((((........)))).....)))))........ (-27.06 = -26.88 + -0.19)

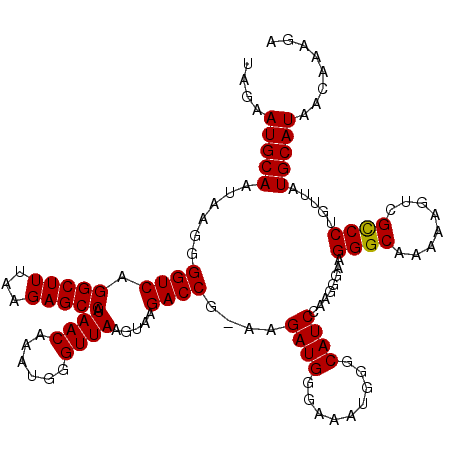
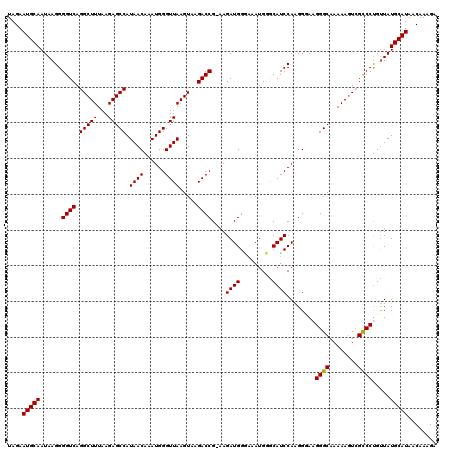
| Location | 7,193,351 – 7,193,470 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.72 |
| Mean single sequence MFE | -24.95 |
| Consensus MFE | -23.21 |
| Energy contribution | -23.15 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7193351 119 - 23771897 UCUUUGUUAUGCAUAACAGGGCGACUUUUUGCCCUUCCCUUGGAUGCCCAUUUUCCAUCUU-CGGUCUUACUUAACCCAUUUGUUAUGGCUCUUAAAGCCUGACCCCUUAUUGCAUUCUA ........(((((....(((((((....))))))).....((((.........))))....-.((((.....((((......)))).((((.....)))).))))......))))).... ( -26.20) >DroSec_CAF1 6847 119 - 1 UCUUUGUUAUGCAUAACGGGGCGACUUUUUGCCCUUCCCUUGGAUGUCCAUUUGCCAUCUU-CGGUCUUACUUAACCCAUUUGUUAUGGCUCUUAAAGCCUGACCCCUUAUUGCAUUCUA ........((((((((((((((((....)))))))..((..(((((.(.....).))))).-.)).................)))))((((.....))))............)))).... ( -26.00) >DroEre_CAF1 6753 120 - 1 UCUUUGUUAUGCAUAACAGGGCGACUUUUUGCCCGUACCCUGGAUGCACAUUCCCCAUCUUCCGGUCUUACUUAACCCAUUUGUUAUGGCUCUUAAAGCCUGACCCCUUAUUGCAUUCUA ....(((((....)))))((((((....)))))).......(((((((...............(((........))).....((((.((((.....)))))))).......))))))).. ( -27.40) >DroYak_CAF1 3848 119 - 1 UCUUUGUUAUGCAUAGCAGGACCAUUUUUUGCCCUUACCCUGGAUGCACAUUCCCCAUCUU-CGGUCUUACUUAACCCAUUUGUUAUGGCUCUUAAAGCCUGACCCCUUAUUGCAUUCUA ........(((((....(((................(((..(((((.........))))).-.)))................((((.((((.....)))))))).)))...))))).... ( -20.20) >consensus UCUUUGUUAUGCAUAACAGGGCGACUUUUUGCCCUUACCCUGGAUGCACAUUCCCCAUCUU_CGGUCUUACUUAACCCAUUUGUUAUGGCUCUUAAAGCCUGACCCCUUAUUGCAUUCUA ....(((((....)))))((((((....)))))).......((((((................(((........))).....((((.((((.....))))))))........)))))).. (-23.21 = -23.15 + -0.06)

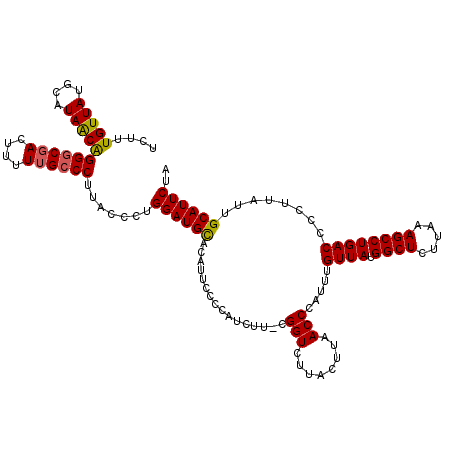
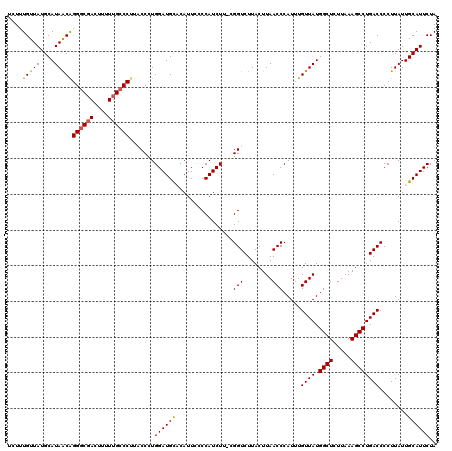
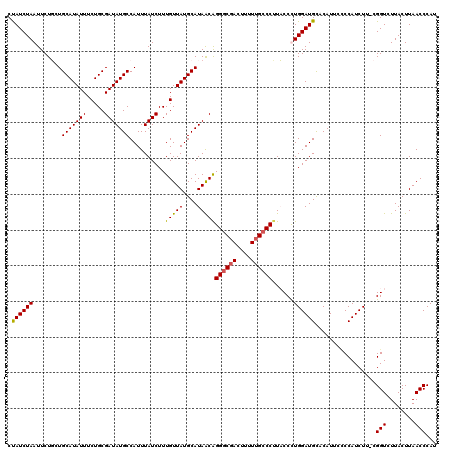
| Location | 7,193,391 – 7,193,510 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -20.90 |
| Consensus MFE | -18.64 |
| Energy contribution | -18.57 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7193391 119 - 23771897 CCAUCUAAUUCUGCUGCAUAUUUCUGCGAUAUGCCAUUUAUCUUUGUUAUGCAUAACAGGGCGACUUUUUGCCCUUCCCUUGGAUGCCCAUUUUCCAUCUU-CGGUCUUACUUAACCCAU .(((((((......((((((...(...((((.......))))...).))))))....(((((((....)))))))....)))))))...............-.(((........)))... ( -23.30) >DroSec_CAF1 6887 119 - 1 CUAUCUAAUUCUGCUGCAUAUUUCUGCGAUAUGCCAUUUAUCUUUGUUAUGCAUAACGGGGCGACUUUUUGCCCUUCCCUUGGAUGUCCAUUUGCCAUCUU-CGGUCUUACUUAACCCAU .(((((((......((((((...(...((((.......))))...).))))))....(((((((....)))))))....))))))).......(((.....-.))).............. ( -22.20) >DroEre_CAF1 6793 120 - 1 CUAUCUAAUUCUUCUGCAUAUUUCUGCGAUAUGCCAUUUAUCUUUGUUAUGCAUAACAGGGCGACUUUUUGCCCGUACCCUGGAUGCACAUUCCCCAUCUUCCGGUCUUACUUAACCCAU ...............(((((((.....)))))))..........(((((....)))))((((((....))))))(.(((..(((((.........)))))...))))............. ( -21.80) >DroYak_CAF1 3888 119 - 1 CUAUCUAAUUCUUCUGCAUAUUUCUGCGAUAUGCCAUUUAUCUUUGUUAUGCAUAGCAGGACCAUUUUUUGCCCUUACCCUGGAUGCACAUUCCCCAUCUU-CGGUCUUACUUAACCCAU ..............((((((...(...((((.......))))...).))))))..((((((.....))))))....(((..(((((.........))))).-.))).............. ( -16.30) >consensus CUAUCUAAUUCUGCUGCAUAUUUCUGCGAUAUGCCAUUUAUCUUUGUUAUGCAUAACAGGGCGACUUUUUGCCCUUACCCUGGAUGCACAUUCCCCAUCUU_CGGUCUUACUUAACCCAU .((((((........(((((((.....)))))))..........(((((....)))))((((((....))))))......)))))).................(((........)))... (-18.64 = -18.57 + -0.06)

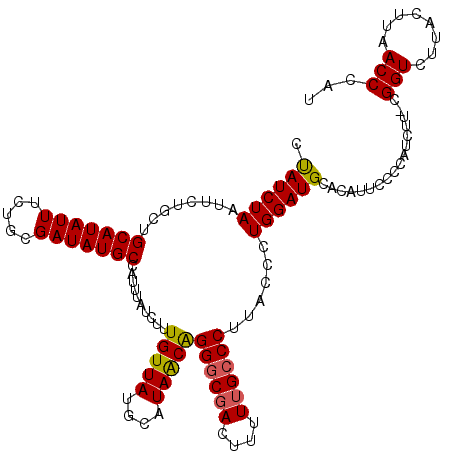
| Location | 7,193,430 – 7,193,550 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -23.89 |
| Energy contribution | -23.95 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7193430 120 + 23771897 AGGGAAGGGCAAAAAGUCGCCCUGUUAUGCAUAACAAAGAUAAAUGGCAUAUCGCAGAAAUAUGCAGCAGAAUUAGAUGGUAAUAGAUGGCAGAAUGCAGCCCCAUCAAAUUCAAGUUAA .....(((((........)))))....(((........((((.......))))(((......))).)))(((((.(((((........(((........)))))))).)))))....... ( -29.10) >DroSec_CAF1 6926 120 + 1 AGGGAAGGGCAAAAAGUCGCCCCGUUAUGCAUAACAAAGAUAAAUGGCAUAUCGCAGAAAUAUGCAGCAGAAUUAGAUAGUAAUAGAUGGCAGAAUGCAGCCCCAUCAAAUUCAAGUUAG .(((..((((........)))).....(((((..((........))((.(((((((......)))............((....))))))))...)))))..)))................ ( -23.30) >DroEre_CAF1 6833 120 + 1 GGGUACGGGCAAAAAGUCGCCCUGUUAUGCAUAACAAAGAUAAAUGGCAUAUCGCAGAAAUAUGCAGAAGAAUUAGAUAGUAAUAGAUGGCAGAAUGCUGCCCUAUCAAACUCAAGUUAA ((((..((((........)))).....((((((.....((((.......)))).......)))))).........(((((........(((((....))))))))))..))))....... ( -27.60) >DroYak_CAF1 3927 120 + 1 GGGUAAGGGCAAAAAAUGGUCCUGCUAUGCAUAACAAAGAUAAAUGGCAUAUCGCAGAAAUAUGCAGAAGAAUUAGAUAGUAAUAGAUGGCAGAAUGCAGCCCUAUCAAAUUCAAGUUAA .((((.((((........)))))))).....((((...........((((((.......))))))....(((((.(((((........(((........)))))))).)))))..)))). ( -25.80) >consensus AGGGAAGGGCAAAAAGUCGCCCUGUUAUGCAUAACAAAGAUAAAUGGCAUAUCGCAGAAAUAUGCAGAAGAAUUAGAUAGUAAUAGAUGGCAGAAUGCAGCCCCAUCAAAUUCAAGUUAA ......((((........)))).........((((...........((((((.......))))))....(((((.(((((........(((........)))))))).)))))..)))). (-23.89 = -23.95 + 0.06)

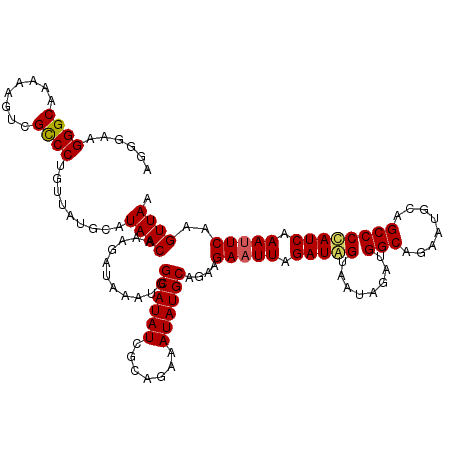
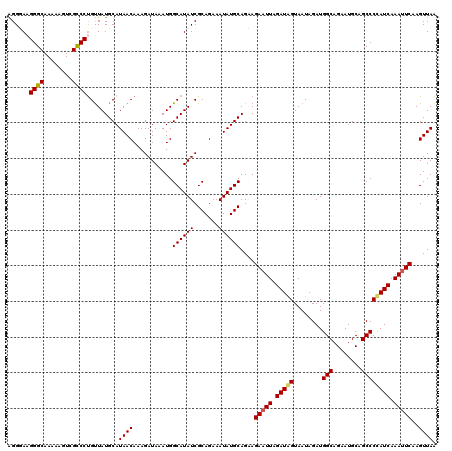
| Location | 7,193,430 – 7,193,550 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -25.35 |
| Energy contribution | -24.85 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7193430 120 - 23771897 UUAACUUGAAUUUGAUGGGGCUGCAUUCUGCCAUCUAUUACCAUCUAAUUCUGCUGCAUAUUUCUGCGAUAUGCCAUUUAUCUUUGUUAUGCAUAACAGGGCGACUUUUUGCCCUUCCCU .((((..(((((.((((((((........)))........))))).)))))....(((((((.....)))))))...........))))........(((((((....)))))))..... ( -28.90) >DroSec_CAF1 6926 120 - 1 CUAACUUGAAUUUGAUGGGGCUGCAUUCUGCCAUCUAUUACUAUCUAAUUCUGCUGCAUAUUUCUGCGAUAUGCCAUUUAUCUUUGUUAUGCAUAACGGGGCGACUUUUUGCCCUUCCCU .......(((((.((((((((........)))........))))).)))))...((((((...(...((((.......))))...).))))))....(((((((....)))))))..... ( -26.60) >DroEre_CAF1 6833 120 - 1 UUAACUUGAGUUUGAUAGGGCAGCAUUCUGCCAUCUAUUACUAUCUAAUUCUUCUGCAUAUUUCUGCGAUAUGCCAUUUAUCUUUGUUAUGCAUAACAGGGCGACUUUUUGCCCGUACCC .......(((((.((((((((((....)))))........))))).)))))....(((((((.....)))))))..........(((((....)))))((((((....))))))...... ( -29.90) >DroYak_CAF1 3927 120 - 1 UUAACUUGAAUUUGAUAGGGCUGCAUUCUGCCAUCUAUUACUAUCUAAUUCUUCUGCAUAUUUCUGCGAUAUGCCAUUUAUCUUUGUUAUGCAUAGCAGGACCAUUUUUUGCCCUUACCC .((((..(((((.((((((((........)))........))))).)))))....(((((((.....)))))))...........))))......((((((.....))))))........ ( -21.10) >consensus UUAACUUGAAUUUGAUAGGGCUGCAUUCUGCCAUCUAUUACUAUCUAAUUCUGCUGCAUAUUUCUGCGAUAUGCCAUUUAUCUUUGUUAUGCAUAACAGGGCGACUUUUUGCCCUUACCC .......(((((.((((((((........)))........))))).)))))....(((((((.....)))))))..........(((((....)))))((((((....))))))...... (-25.35 = -24.85 + -0.50)

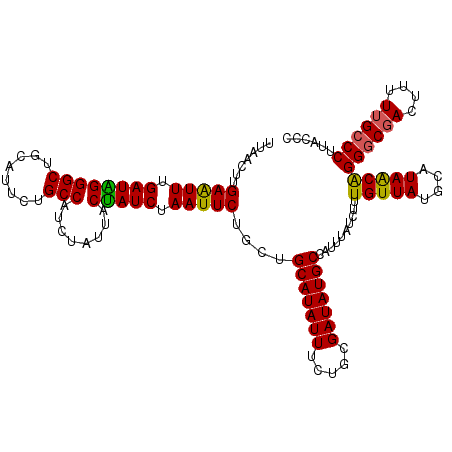

| Location | 7,193,470 – 7,193,590 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.86 |
| Mean single sequence MFE | -28.66 |
| Consensus MFE | -23.89 |
| Energy contribution | -24.70 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7193470 120 - 23771897 UUUAACGGGUUAUUUUGCUUGAGUGCGAGCCAUCAAAUAUUUAACUUGAAUUUGAUGGGGCUGCAUUCUGCCAUCUAUUACCAUCUAAUUCUGCUGCAUAUUUCUGCGAUAUGCCAUUUA .......(((......((..(((((((..(((((((((...........)))))))))..).)))))).))........))).............(((((((.....)))))))...... ( -27.64) >DroSec_CAF1 6966 120 - 1 UUUAACUGGUUAUUUUGCUUGAGUGCGGGCCAUCAAAUAUCUAACUUGAAUUUGAUGGGGCUGCAUUCUGCCAUCUAUUACUAUCUAAUUCUGCUGCAUAUUUCUGCGAUAUGCCAUUUA ......((((....((((..((((((((.(((((((((.((......)))))))))))(((........))).....................))))))...)).))))...)))).... ( -32.10) >DroEre_CAF1 6873 120 - 1 UUUAACGGGUGCUUUUGCUUGAGUGAGAGCCAUCAAAUAUUUAACUUGAGUUUGAUAGGGCAGCAUUCUGCCAUCUAUUACUAUCUAAUUCUUCUGCAUAUUUCUGCGAUAUGCCAUUUA .......((((((((..(....)..)))).)))).............(((((.((((((((((....)))))........))))).)))))....(((((((.....)))))))...... ( -29.30) >DroYak_CAF1 3967 120 - 1 UUUAACGGGUGAUUUUGCUUGAGUGAGAGCCAUCAAAUAUUUAACUUGAAUUUGAUAGGGCUGCAUUCUGCCAUCUAUUACUAUCUAAUUCUUCUGCAUAUUUCUGCGAUAUGCCAUUUA .......((((((...((..(((((..(((((((((((...........)))))))..)))).))))).)).....)))))).............(((((((.....)))))))...... ( -25.60) >consensus UUUAACGGGUGAUUUUGCUUGAGUGAGAGCCAUCAAAUAUUUAACUUGAAUUUGAUAGGGCUGCAUUCUGCCAUCUAUUACUAUCUAAUUCUGCUGCAUAUUUCUGCGAUAUGCCAUUUA .......((((((...((..((((((.(((((((((((.((......)))))))))..)))))))))).)).....)))))).............(((((((.....)))))))...... (-23.89 = -24.70 + 0.81)

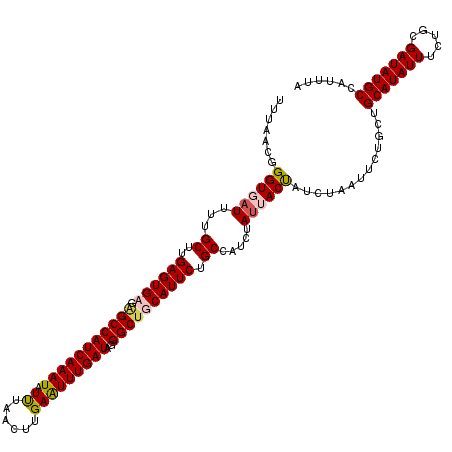
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:11 2006