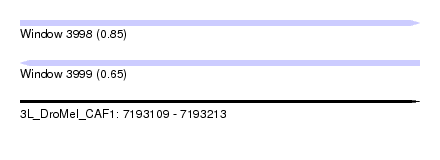
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,193,109 – 7,193,213 |
| Length | 104 |
| Max. P | 0.848800 |

| Location | 7,193,109 – 7,193,213 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 107 |
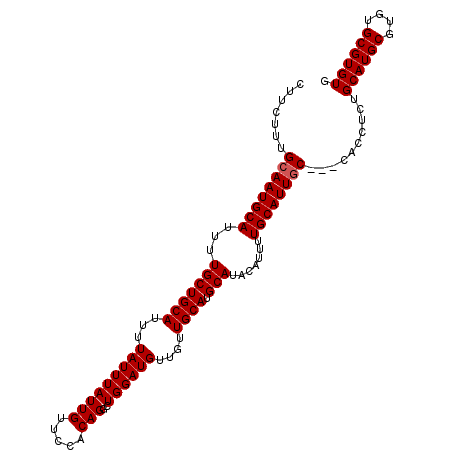
| Reading direction | forward |
| Mean pairwise identity | 97.63 |
| Mean single sequence MFE | -24.68 |
| Consensus MFE | -22.05 |
| Energy contribution | -22.30 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7193109 104 + 23771897 CACACGCACACGCAUGCAGAGGUG---GCAAUGCAAAAUGUAUGCAUGCAACAACAUCCAGCCUGUGGAACAAUAAAUAAAAUGCAGCAAAAUGCAUUUCAAAGAAG ((((.((....(((((((...((.---.....))....)))))))(((......)))...)).))))............(((((((......)))))))........ ( -23.00) >DroSec_CAF1 6598 107 + 1 CACACGCACACGCAUGCAGAGGUGGCGGCAAUGCAAAAUGUAUGCAUGCAACAACAUCCAGCCUGUGGAACAAUAAAUAAAAUGCAGCAAAAUGCAUUGCAAAGAAG ....(((.(((..........))))))((((((((...(((.(((((.........((((.....))))............))))))))...))))))))....... ( -27.30) >DroEre_CAF1 6523 104 + 1 CACACGCACACGCAUGCAGAGGUG---GCAAUGCAAAAUGUAUGCAUGCAACAACAUCCAGCCUGUGGAACAAUAAAUAAAAUGCAGCAAAAUGCAUUGCAAAGAAG (((..(((......)))....)))---((((((((...(((.(((((.........((((.....))))............))))))))...))))))))....... ( -24.70) >DroYak_CAF1 3606 104 + 1 UACACGCACACGCAUGCAGAGGUG---GCAAUGCAAAAUGUAUGCAUGCAACAACAUCCAGCCUGUGGAACAAUAAAUAAAAUGCAGCAAAAUGCAUUGCAAAGAAG .....((....(((((((.(.((.---((...))...)).).))))))).......((((.....))))...........((((((......))))))))....... ( -23.70) >consensus CACACGCACACGCAUGCAGAGGUG___GCAAUGCAAAAUGUAUGCAUGCAACAACAUCCAGCCUGUGGAACAAUAAAUAAAAUGCAGCAAAAUGCAUUGCAAAGAAG .....((...((((.((.(.((((...(((.((((.......))))))).....))))).)).)))).............((((((......))))))))....... (-22.05 = -22.30 + 0.25)

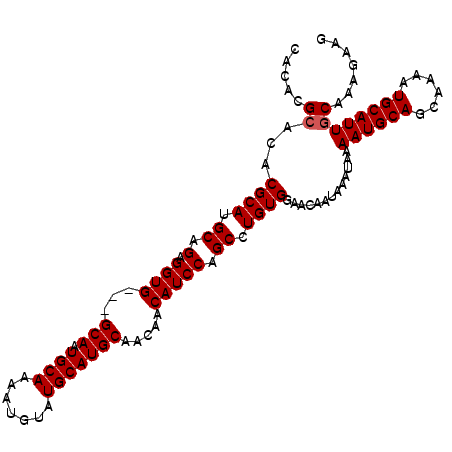
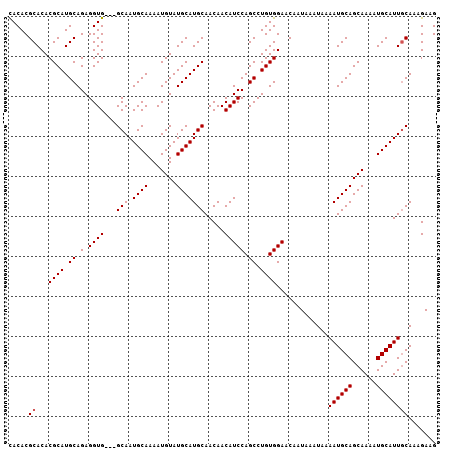
| Location | 7,193,109 – 7,193,213 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 97.63 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -24.67 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7193109 104 - 23771897 CUUCUUUGAAAUGCAUUUUGCUGCAUUUUAUUUAUUGUUCCACAGGCUGGAUGUUGUUGCAUGCAUACAUUUUGCAUUGC---CACCUCUGCAUGCGUGUGCGUGUG ......(((((((((......)))))))))........((((.....)))).......((((((((((.....((((.((---.......)))))))))))))))). ( -27.80) >DroSec_CAF1 6598 107 - 1 CUUCUUUGCAAUGCAUUUUGCUGCAUUUUAUUUAUUGUUCCACAGGCUGGAUGUUGUUGCAUGCAUACAUUUUGCAUUGCCGCCACCUCUGCAUGCGUGUGCGUGUG .......((((((((...(((((((...(((((((((.....)))..))))))....)))).))).......))))))))..........((((((....)))))). ( -26.90) >DroEre_CAF1 6523 104 - 1 CUUCUUUGCAAUGCAUUUUGCUGCAUUUUAUUUAUUGUUCCACAGGCUGGAUGUUGUUGCAUGCAUACAUUUUGCAUUGC---CACCUCUGCAUGCGUGUGCGUGUG .......((((((((...(((((((...(((((((((.....)))..))))))....)))).))).......))))))))---.......((((((....)))))). ( -26.90) >DroYak_CAF1 3606 104 - 1 CUUCUUUGCAAUGCAUUUUGCUGCAUUUUAUUUAUUGUUCCACAGGCUGGAUGUUGUUGCAUGCAUACAUUUUGCAUUGC---CACCUCUGCAUGCGUGUGCGUGUA .......((((((((...(((((((...(((((((((.....)))..))))))....)))).))).......))))))))---......(((((((....))))))) ( -27.10) >consensus CUUCUUUGCAAUGCAUUUUGCUGCAUUUUAUUUAUUGUUCCACAGGCUGGAUGUUGUUGCAUGCAUACAUUUUGCAUUGC___CACCUCUGCAUGCGUGUGCGUGUG .......((((((((...(((((((...(((((((((.....)))..))))))....)))).))).......))))))))..........((((((....)))))). (-24.67 = -24.92 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:04 2006