
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
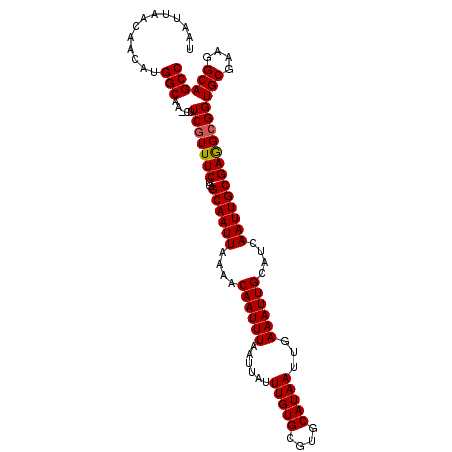
| Location | 7,192,760 – 7,192,865 |
| Length | 105 |
| Max. P | 0.967636 |

| Location | 7,192,760 – 7,192,865 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 96.82 |
| Mean single sequence MFE | -25.78 |
| Consensus MFE | -24.99 |
| Energy contribution | -25.05 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7192760 105 + 23771897 UAAUUAACAACAUGGCAAAUUUCGUUUCUGGCAAUUAAAACAAUUUAAUUAUUUGUGCGUGCAUAAUUGAAAUUGGAUCAAUUGCGAGGCGGUGCGAAGGCAGCC .............(((.....(((((((..((((((.(..((((((......(((((....)))))...))))))..).)))))))))))))(((....)))))) ( -27.10) >DroSec_CAF1 6259 105 + 1 UAAUUAACAACAUGGCAAAUUUCAUUUCUGGCAAUUAAAACAAUUUAAUUAUUUGUGCGUGCAUAAUUGAAAUUGCAUCAAUUGCGAGGCGGUGCGAAGGCAGCC .............(((..........(((.((((((....((((((......(((((....)))))...))))))....)))))).)))...(((....)))))) ( -23.30) >DroEre_CAF1 6179 104 + 1 UAAUUAACAACAUGGCAA-CUUCGUUUCUGGCAAUUAAAACAAUUUAAUUAUUUGUGCGUGCAUAAUUGAAAUUGCAUCAAUUGCGAGGCGGUGCGAAGGCAGCC .............(((..-((((((.(((.((((((....((((((......(((((....)))))...))))))....)))))).)))....))))))...))) ( -26.40) >DroYak_CAF1 3239 104 + 1 UAAUUAACGACAUGGCAA-CUUCGUUUCUGGCAAUUAAAACAAUUUAAUUAUUUGUGCGUGCAUAAUUGAAAUUGCAUCAAUUGCGAAGCGGUGCGAAGGCAGCC .............(((..-..(((((((..((((((....((((((......(((((....)))))...))))))....)))))))))))))(((....)))))) ( -26.30) >consensus UAAUUAACAACAUGGCAA_CUUCGUUUCUGGCAAUUAAAACAAUUUAAUUAUUUGUGCGUGCAUAAUUGAAAUUGCAUCAAUUGCGAGGCGGUGCGAAGGCAGCC .............(((.....(((((((..((((((....((((((......(((((....)))))...))))))....)))))))))))))(((....)))))) (-24.99 = -25.05 + 0.06)


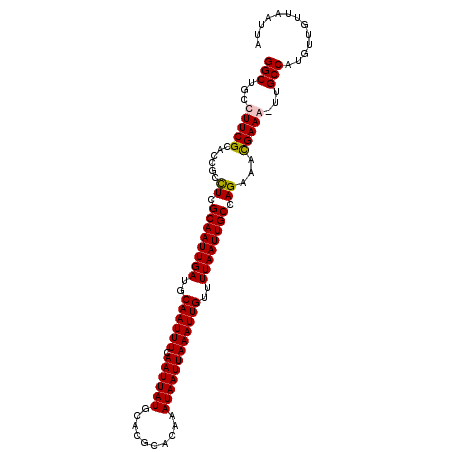
| Location | 7,192,760 – 7,192,865 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 96.82 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -23.98 |
| Energy contribution | -24.10 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7192760 105 - 23771897 GGCUGCCUUCGCACCGCCUCGCAAUUGAUCCAAUUUCAAUUAUGCACGCACAAAUAAUUAAAUUGUUUUAAUUGCCAGAAACGAAAUUUGCCAUGUUGUUAAUUA (((....((((......((.((((((((..((((((.((((((..........))))))))))))..)))))))).))...))))....)))............. ( -23.40) >DroSec_CAF1 6259 105 - 1 GGCUGCCUUCGCACCGCCUCGCAAUUGAUGCAAUUUCAAUUAUGCACGCACAAAUAAUUAAAUUGUUUUAAUUGCCAGAAAUGAAAUUUGCCAUGUUGUUAAUUA ((((((....)))....((.((((((((.(((((((.((((((..........))))))))))))).)))))))).))...........)))............. ( -23.40) >DroEre_CAF1 6179 104 - 1 GGCUGCCUUCGCACCGCCUCGCAAUUGAUGCAAUUUCAAUUAUGCACGCACAAAUAAUUAAAUUGUUUUAAUUGCCAGAAACGAAG-UUGCCAUGUUGUUAAUUA (((...(((((......((.((((((((.(((((((.((((((..........))))))))))))).)))))))).))...)))))-..)))............. ( -26.80) >DroYak_CAF1 3239 104 - 1 GGCUGCCUUCGCACCGCUUCGCAAUUGAUGCAAUUUCAAUUAUGCACGCACAAAUAAUUAAAUUGUUUUAAUUGCCAGAAACGAAG-UUGCCAUGUCGUUAAUUA (((...(((((.....((..((((((((.(((((((.((((((..........))))))))))))).)))))))).))...)))))-..)))............. ( -26.90) >consensus GGCUGCCUUCGCACCGCCUCGCAAUUGAUGCAAUUUCAAUUAUGCACGCACAAAUAAUUAAAUUGUUUUAAUUGCCAGAAACGAAA_UUGCCAUGUUGUUAAUUA (((...(((((......((.((((((((..((((((.((((((..........))))))))))))..)))))))).))...)))))...)))............. (-23.98 = -24.10 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:03 2006