
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,192,365 – 7,192,507 |
| Length | 142 |
| Max. P | 0.964840 |

| Location | 7,192,365 – 7,192,467 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 96.73 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -26.47 |
| Energy contribution | -26.03 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
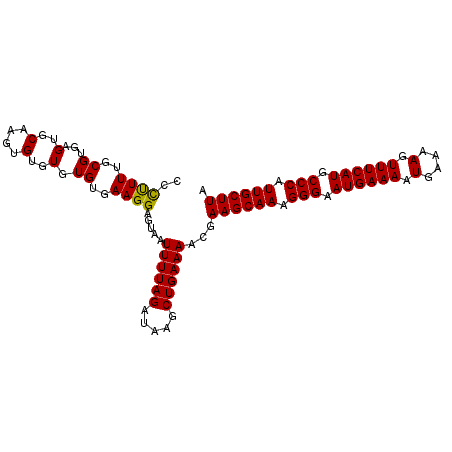
>3L_DroMel_CAF1 7192365 102 + 23771897 CCCCUUUGCGUAAGUGCAAGUGUGUGUGUGAAGGAGUAAUUUUAGAUAAGCUGAAAACGAAGCAAAGGGAAUGAAAAUGAAAAGUUUCAUGCCCAUUGCUUA ..((((..((...(..(....)..).))..)))).....((((((.....))))))...((((((.(((.((((((.(....).)))))).))).)))))). ( -29.60) >DroSec_CAF1 5857 102 + 1 CCCUUUUGCGUGAGUGCAAGUGUGUGUGUGAAGGAGUAAUUUUAGAUAAGCUGAAAACGAAGCAAAGGGAAUGAAAAUGAAAAGUUUCAUGCCCAUUGCUUA ..((((..((...(..(....)..).))..)))).....((((((.....))))))...((((((.(((.((((((.(....).)))))).))).)))))). ( -26.90) >DroEre_CAF1 5798 102 + 1 CCUUUUUGCGUGAGUGCAAGUGUGUGUGUGAAGGAGUAAUUUUAGAUAAGCUGAAAACGAAGCAAAGGGAAUGAAAUUGAAAACUUUCAUGCCCAUUGCUUA (((((..((....))(((......)))..))))).....((((((.....))))))...((((((.(((.((((((.(....).)))))).))).)))))). ( -26.20) >consensus CCCUUUUGCGUGAGUGCAAGUGUGUGUGUGAAGGAGUAAUUUUAGAUAAGCUGAAAACGAAGCAAAGGGAAUGAAAAUGAAAAGUUUCAUGCCCAUUGCUUA ..((((..((...(..(....)..).))..)))).....((((((.....))))))...((((((.(((.((((((........)))))).))).)))))). (-26.47 = -26.03 + -0.44)

| Location | 7,192,365 – 7,192,467 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 96.73 |
| Mean single sequence MFE | -20.23 |
| Consensus MFE | -18.80 |
| Energy contribution | -18.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
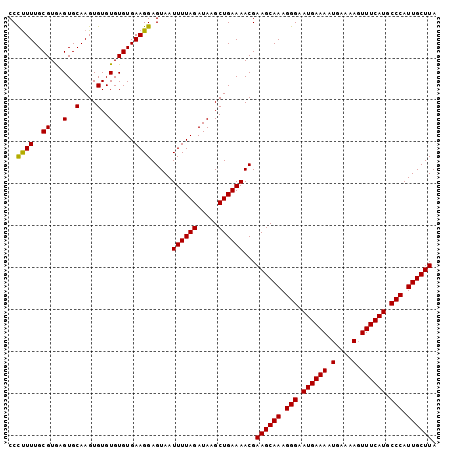
>3L_DroMel_CAF1 7192365 102 - 23771897 UAAGCAAUGGGCAUGAAACUUUUCAUUUUCAUUCCCUUUGCUUCGUUUUCAGCUUAUCUAAAAUUACUCCUUCACACACACACUUGCACUUACGCAAAGGGG .((((((.(((.((((((........)))))).))).)))))).......................((((((............(((......))))))))) ( -21.50) >DroSec_CAF1 5857 102 - 1 UAAGCAAUGGGCAUGAAACUUUUCAUUUUCAUUCCCUUUGCUUCGUUUUCAGCUUAUCUAAAAUUACUCCUUCACACACACACUUGCACUCACGCAAAAGGG .((((((.(((.((((((........)))))).))).)))))).........................((((...........((((......)))))))). ( -19.80) >DroEre_CAF1 5798 102 - 1 UAAGCAAUGGGCAUGAAAGUUUUCAAUUUCAUUCCCUUUGCUUCGUUUUCAGCUUAUCUAAAAUUACUCCUUCACACACACACUUGCACUCACGCAAAAAGG .((((((.(((.((((((........)))))).))).))))))........................................((((......))))..... ( -19.40) >consensus UAAGCAAUGGGCAUGAAACUUUUCAUUUUCAUUCCCUUUGCUUCGUUUUCAGCUUAUCUAAAAUUACUCCUUCACACACACACUUGCACUCACGCAAAAGGG .((((((.(((.((((((........)))))).))).))))))........................................((((......))))..... (-18.80 = -18.80 + 0.00)



| Location | 7,192,393 – 7,192,507 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.45 |
| Mean single sequence MFE | -25.71 |
| Consensus MFE | -21.89 |
| Energy contribution | -21.77 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
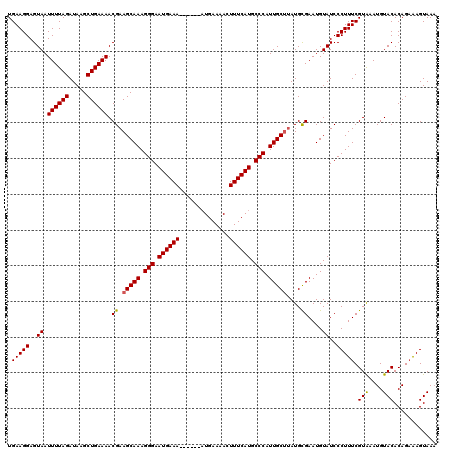
>3L_DroMel_CAF1 7192393 114 + 23771897 UGAAGGAGUAAUUUUAGAUAAGCUGAAAACGAAGCAAAGGGAAUGAAA------AUGAAAAGUUUCAUGCCCAUUGCUUAUGCGAAUGUAUCCCUUUCGUAAAUGUACACAGAAAGUAAA .(((((..((.((((((.....))))))...((((((.(((.((((((------.(....).)))))).))).)))))).........))..))))).(((....)))............ ( -25.30) >DroSec_CAF1 5885 114 + 1 UGAAGGAGUAAUUUUAGAUAAGCUGAAAACGAAGCAAAGGGAAUGAAA------AUGAAAAGUUUCAUGCCCAUUGCUUAUGUGAAUGUAUCCCUUUCGUAAAUGUACACAGAGAGUAAC ....(((....((((((.....))))))(((((((((.(((.((((((------.(....).)))))).))).)))))).))).......)))(((((((........)).))))).... ( -26.40) >DroEre_CAF1 5826 114 + 1 UGAAGGAGUAAUUUUAGAUAAGCUGAAAACGAAGCAAAGGGAAUGAAA------UUGAAAACUUUCAUGCCCAUUGCUUAUGCGAAUGUAUCCCUUUCGUAAAUGUACACAGAAAGUAAA .(((((..((.((((((.....))))))...((((((.(((.((((((------.(....).)))))).))).)))))).........))..))))).(((....)))............ ( -25.30) >DroYak_CAF1 2878 120 + 1 UGAAGGAGUAAUUUUAGAUAAGCUGAAAACGAAGCAAAGGGAAUGAAAAUGAAAAUGAAAACUUUCAUGCCCAUUGCAGAUGCGAAUGUAUCCCUUUCGUGAAUGUACACAGCAAGUAAA .(((((.....((((((.....)))))).....((((.(((.((((((..............)))))).))).)))).(((((....))))).)))))(((......))).......... ( -25.84) >consensus UGAAGGAGUAAUUUUAGAUAAGCUGAAAACGAAGCAAAGGGAAUGAAA______AUGAAAACUUUCAUGCCCAUUGCUUAUGCGAAUGUAUCCCUUUCGUAAAUGUACACAGAAAGUAAA .(((((..((.((((((.....)))))).((.(((((.(((.((((((..............)))))).))).)))))....))....))..))))).(((....)))............ (-21.89 = -21.77 + -0.12)


| Location | 7,192,393 – 7,192,507 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.45 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -19.25 |
| Energy contribution | -20.07 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
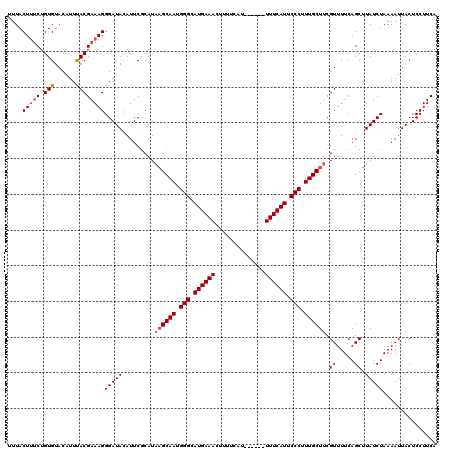
>3L_DroMel_CAF1 7192393 114 - 23771897 UUUACUUUCUGUGUACAUUUACGAAAGGGAUACAUUCGCAUAAGCAAUGGGCAUGAAACUUUUCAU------UUUCAUUCCCUUUGCUUCGUUUUCAGCUUAUCUAAAAUUACUCCUUCA ....(((((.(((......))))))))(((((.........((((((.(((.((((((........------)))))).))).)))))).((.....)).)))))............... ( -23.30) >DroSec_CAF1 5885 114 - 1 GUUACUCUCUGUGUACAUUUACGAAAGGGAUACAUUCACAUAAGCAAUGGGCAUGAAACUUUUCAU------UUUCAUUCCCUUUGCUUCGUUUUCAGCUUAUCUAAAAUUACUCCUUCA ((...(((((.((((....))))..))))).))........((((((.(((.((((((........------)))))).))).))))))............................... ( -20.50) >DroEre_CAF1 5826 114 - 1 UUUACUUUCUGUGUACAUUUACGAAAGGGAUACAUUCGCAUAAGCAAUGGGCAUGAAAGUUUUCAA------UUUCAUUCCCUUUGCUUCGUUUUCAGCUUAUCUAAAAUUACUCCUUCA ....(((((.(((......))))))))(((((.........((((((.(((.((((((........------)))))).))).)))))).((.....)).)))))............... ( -24.20) >DroYak_CAF1 2878 120 - 1 UUUACUUGCUGUGUACAUUCACGAAAGGGAUACAUUCGCAUCUGCAAUGGGCAUGAAAGUUUUCAUUUUCAUUUUCAUUCCCUUUGCUUCGUUUUCAGCUUAUCUAAAAUUACUCCUUCA .......((((...((((((.(....)))))............((((.(((.((((((((..........)))))))).))).))))...))...))))..................... ( -25.60) >consensus UUUACUUUCUGUGUACAUUUACGAAAGGGAUACAUUCGCAUAAGCAAUGGGCAUGAAACUUUUCAU______UUUCAUUCCCUUUGCUUCGUUUUCAGCUUAUCUAAAAUUACUCCUUCA ....(((((.(((......))))))))(((((.........((((((.(((.((((((..............)))))).))).)))))).((.....)).)))))............... (-19.25 = -20.07 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:01 2006