
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,190,106 – 7,190,294 |
| Length | 188 |
| Max. P | 0.780574 |

| Location | 7,190,106 – 7,190,226 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -35.77 |
| Consensus MFE | -29.58 |
| Energy contribution | -29.82 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7190106 120 - 23771897 UGGACCGAAAACCCGUUGAGACUCAUAAAAAUGUGCCACUUAGCAGGGAAGUUGGGUGGCCAACGGGGGAAUUGGUUAAAAGGAGGAGCAAUGGAGGCUGGUGGAUUGAUAGGGGGUGGC ..((((((...(((((((.....(((....))).((((((((((......)))))))))))))))))....)))))).........(((.......)))..................... ( -33.80) >DroSec_CAF1 3562 120 - 1 UGGACCGAAAACCCGUUGAGACUCAUAAAAAUGUGCCACUUAGCAGGGAAGUUGGGUGGCCAACCGGGGAAUUGCUUAAAAGGAGGAGCAAUGGAGGCUGGUGGAUUGAUAGGGGGUGGC ....(((....(((((..(..(.........((.((((((((((......))))))))))))(((((...(((((((........))))))).....))))))..)..)).)))..))). ( -38.80) >DroEre_CAF1 3478 119 - 1 UGGAGCGAAAACCCGUUGAGACUCAUAAAAAUGUGCCACUUAGCAGGGAAGUUGGGUGGCUAGCCGGGGAAUUGGUUAAAAGAAGGAGCGAUGGAGGCUGGUGGAUUGAUAGGG-GUGGC ....((....((((.(...((..(((........((((((((((......))))))))))(((((.....((((.((........)).))))...))))))))..))...).))-)).)) ( -35.10) >DroYak_CAF1 479 120 - 1 UGGACCGAAAACCCGUUGAGACUCAUAAAAAUGUGCCACUUAGCAGGGAAGUUGGGUGGCCAACCGGGGAAUUGGUUAAAAGAAGGAGCCAUGGAAGCUGGUGGAUUGAUAGGGGGUGGC ....(((....(((((..(..(.........((.((((((((((......))))))))))))(((((.....(((((.........)))))......))))))..)..)).)))..))). ( -35.40) >consensus UGGACCGAAAACCCGUUGAGACUCAUAAAAAUGUGCCACUUAGCAGGGAAGUUGGGUGGCCAACCGGGGAAUUGGUUAAAAGAAGGAGCAAUGGAGGCUGGUGGAUUGAUAGGGGGUGGC ....(((....(((((..(..(.........((.((((((((((......))))))))))))(((((...((((.((........)).)))).....))))))..)..)).)))..))). (-29.58 = -29.82 + 0.25)


| Location | 7,190,146 – 7,190,254 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.38 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -25.80 |
| Energy contribution | -25.93 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7190146 108 - 23771897 GCCAGA------------GAAGCAGCAGAAAUUAUCAACUUGGACCGAAAACCCGUUGAGACUCAUAAAAAUGUGCCACUUAGCAGGGAAGUUGGGUGGCCAACGGGGGAAUUGGUUAAA ((....------------...))...................((((((...(((((((.....(((....))).((((((((((......)))))))))))))))))....))))))... ( -32.40) >DroSec_CAF1 3602 108 - 1 GCCAGA------------GAAGCAGCAGAAAUUAUCAACUUGGACCGAAAACCCGUUGAGACUCAUAAAAAUGUGCCACUUAGCAGGGAAGUUGGGUGGCCAACCGGGGAAUUGCUUAAA ......------------.((((((.........(((((..((........)).)))))..(((.......((.((((((((((......))))))))))))....)))..))))))... ( -27.10) >DroEre_CAF1 3517 120 - 1 GCCAGUGAAACAGCAGCAGAAGCAGCAGAAAUUAUCAACUUGGAGCGAAAACCCGUUGAGACUCAUAAAAAUGUGCCACUUAGCAGGGAAGUUGGGUGGCUAGCCGGGGAAUUGGUUAAA ((((((......((.((....)).))............(((((((((......)))).................((((((((((......))))))))))...)))))..)))))).... ( -31.90) >DroYak_CAF1 519 114 - 1 GCCAGAGAAACA------GAAGCAGCAGAAAUUAUCAACUUGGACCGAAAACCCGUUGAGACUCAUAAAAAUGUGCCACUUAGCAGGGAAGUUGGGUGGCCAACCGGGGAAUUGGUUAAA ............------........................((((((...(((((((.....(((....))).((((((((((......))))))))))))).))))...))))))... ( -29.10) >consensus GCCAGA____________GAAGCAGCAGAAAUUAUCAACUUGGACCGAAAACCCGUUGAGACUCAUAAAAAUGUGCCACUUAGCAGGGAAGUUGGGUGGCCAACCGGGGAAUUGGUUAAA ..........................................((((((...(((((((.....(((....))).((((((((((......)))))))))))))).)))...))))))... (-25.80 = -25.93 + 0.13)


| Location | 7,190,186 – 7,190,294 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -26.16 |
| Energy contribution | -26.16 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
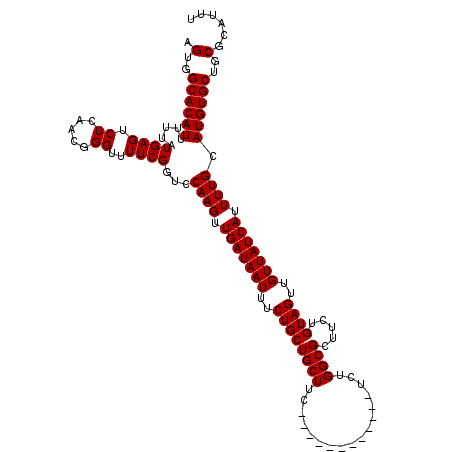
>3L_DroMel_CAF1 7190186 108 + 23771897 AGUGGCACAUUUUUAUGAGUCUCAACGGGUUUUCGGUCCAAGUUGAUAAUUUCUGCUGCUUC------------UCUGGCGCUUCUGUAGUUGUUAUCAUUUUGCAUGUGCUGCGCAUUU .(..((((((.....((((.((.....))..))))...((((.(((((((..((((((((..------------...)))).....))))..))))))).)))).))))))..)...... ( -28.40) >DroSec_CAF1 3642 108 + 1 AGUGGCACAUUUUUAUGAGUCUCAACGGGUUUUCGGUCCAAGUUGAUAAUUUCUGCUGCUUC------------UCUGGCGCUUCUGUAGUUGUUAUCAUUUUGCAUGUGCUGCGCAUUU .(..((((((.....((((.((.....))..))))...((((.(((((((..((((((((..------------...)))).....))))..))))))).)))).))))))..)...... ( -28.40) >DroEre_CAF1 3557 120 + 1 AGUGGCACAUUUUUAUGAGUCUCAACGGGUUUUCGCUCCAAGUUGAUAAUUUCUGCUGCUUCUGCUGCUGUUUCACUGGCGCUUCUGUAGUUGUUAUCAUUUUGCAUGUGCUCCGCAUUU .(..((((((................((((....))))((((.(((((((....(((((....((.(((........)))))....))))).))))))).)))).))))))..)...... ( -29.70) >DroYak_CAF1 559 114 + 1 AGUGGCACAUUUUUAUGAGUCUCAACGGGUUUUCGGUCCAAGUUGAUAAUUUCUGCUGCUUC------UGUUUCUCUGGCGCUUCUGUAGUUGUUAUCAUUUUGCAUGUGCUGCGCAUUU .(..((((((.....((((.((.....))..))))...((((.(((((((..((((.((.((------.........)).))....))))..))))))).)))).))))))..)...... ( -28.30) >consensus AGUGGCACAUUUUUAUGAGUCUCAACGGGUUUUCGGUCCAAGUUGAUAAUUUCUGCUGCUUC____________UCUGGCGCUUCUGUAGUUGUUAUCAUUUUGCAUGUGCUGCGCAUUU .(..((((((.....((((.((.....))..))))...((((.(((((((..((((((((.................)))).....))))..))))))).)))).))))))..)...... (-26.16 = -26.16 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:57 2006