| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,187,682 – 7,187,817 |
| Length | 135 |
| Max. P | 0.994100 |

| Location | 7,187,682 – 7,187,777 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 84.08 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -17.60 |
| Energy contribution | -19.00 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
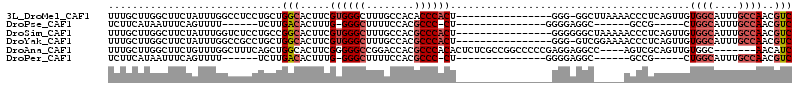
>3L_DroMel_CAF1 7187682 95 + 23771897 AACAAUAAAACUUCCGCUUU-GGCGUUGAAGUUGUAGUUGACGUUGGCAAAUGCCACAACUGAGGGUUUUAAGCC-CCC-AGUGGGUGUGGCAAAGCC ..((((..(((((((((...-.)))..))))))...)))).....(((...(((((((((((.((((.....)))-).)-)))...)))))))..))) ( -36.00) >DroPse_CAF1 54653 86 + 1 AACAAUAAAACUUCAGCUUUUGCCGUUGAUGUUGAAGUUGACGUUGGCAAAUGCCAG-----CGGC------GCCUCCCCAG-GGGCGUGGAAAAGCC ...............((((((.((((..((......))..))((((((....)))))-----).((------(((((....)-)))))))))))))). ( -33.00) >DroSim_CAF1 51177 96 + 1 AACAAUAAAACUUCCGCUUU-GGCGUUGAAGUUGUAGUUGACGUUGGCAAAUGCCACAACUGAGGGUUUUUAGCCCCCC-AGUGGGCGUGGCAAAGCC ..((((..(((((((((...-.)))..))))))...)))).....(((...((((((.((((.((((.....))))..)-)))....))))))..))) ( -33.80) >DroYak_CAF1 54832 95 + 1 AACAAUAAAACUUCCGCUUU-GCCGUUGAAGUUGUAGUUGACGUUGGCAAAUGCCACAACUGAGGGUUUUCCGAC-CCC-AGUGGGCGUGGCAAAGCC ...............(((((-(((((..(.(((......))).)..))..(((((...((((.(((((....)))-)))-))).))))))))))))). ( -38.70) >DroPer_CAF1 63761 86 + 1 AACAAUAAAACUUCAGCUUUUGCCGUUGAUGUUGAAGUUGACGUUGGCAAAUGCCAG-----CGGC------GCCUCCCCAG-GGGCGUGGAAAAGCC ...............((((((.((((..((......))..))((((((....)))))-----).((------(((((....)-)))))))))))))). ( -33.00) >consensus AACAAUAAAACUUCCGCUUU_GCCGUUGAAGUUGUAGUUGACGUUGGCAAAUGCCACAACUGAGGGUUUU__GCC_CCC_AGUGGGCGUGGCAAAGCC ..((((..((((((.((.......)).))))))...)))).....(((...((((((......((........)).(((....))).))))))..))) (-17.60 = -19.00 + 1.40)

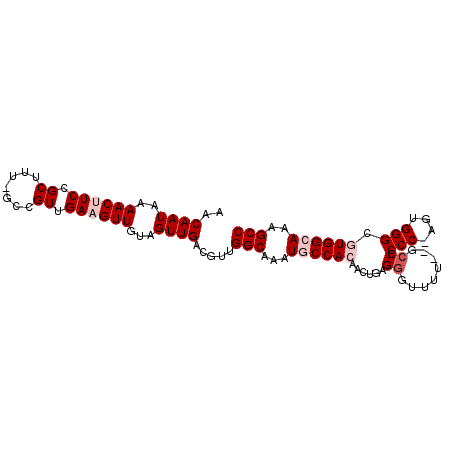
| Location | 7,187,682 – 7,187,777 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 84.08 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -18.42 |
| Energy contribution | -19.42 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7187682 95 - 23771897 GGCUUUGCCACACCCACU-GGG-GGCUUAAAACCCUCAGUUGUGGCAUUUGCCAACGUCAACUACAACUUCAACGCC-AAAGCGGAAGUUUUAUUGUU (((..(((((((...(((-(((-((.......)))))))))))))))...))).....(((.((.((((((..(((.-...))))))))).))))).. ( -37.10) >DroPse_CAF1 54653 86 - 1 GGCUUUUCCACGCCC-CUGGGGAGGC------GCCG-----CUGGCAUUUGCCAACGUCAACUUCAACAUCAACGGCAAAAGCUGAAGUUUUAUUGUU .((((..(((.....-.)))..))))------((((-----.((((....))))..((........)).....)))).(((((....)))))...... ( -24.60) >DroSim_CAF1 51177 96 - 1 GGCUUUGCCACGCCCACU-GGGGGGCUAAAAACCCUCAGUUGUGGCAUUUGCCAACGUCAACUACAACUUCAACGCC-AAAGCGGAAGUUUUAUUGUU (((..(((((((...(((-(((((........)))))))))))))))...))).....(((.((.((((((..(((.-...))))))))).))))).. ( -38.00) >DroYak_CAF1 54832 95 - 1 GGCUUUGCCACGCCCACU-GGG-GUCGGAAAACCCUCAGUUGUGGCAUUUGCCAACGUCAACUACAACUUCAACGGC-AAAGCGGAAGUUUUAUUGUU (((..(((((((...(((-(((-((......)))).)))))))))))...)))..................(((((.-(((((....))))).))))) ( -34.20) >DroPer_CAF1 63761 86 - 1 GGCUUUUCCACGCCC-CUGGGGAGGC------GCCG-----CUGGCAUUUGCCAACGUCAACUUCAACAUCAACGGCAAAAGCUGAAGUUUUAUUGUU .((((..(((.....-.)))..))))------((((-----.((((....))))..((........)).....)))).(((((....)))))...... ( -24.60) >consensus GGCUUUGCCACGCCCACU_GGG_GGC__AAAACCCUCAGUUGUGGCAUUUGCCAACGUCAACUACAACUUCAACGGC_AAAGCGGAAGUUUUAUUGUU (((..((((((.(((....))).((........))......))))))...)))..................((((...(((((....)))))..)))) (-18.42 = -19.42 + 1.00)

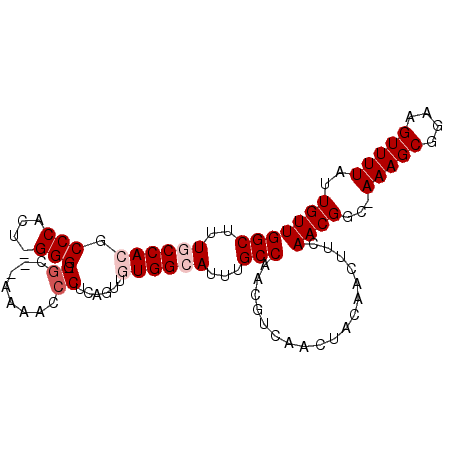
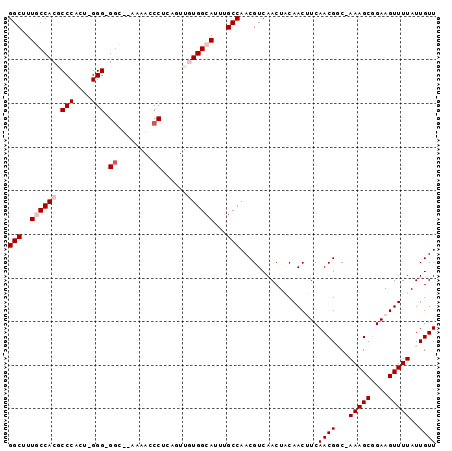
| Location | 7,187,720 – 7,187,817 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 66.91 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -16.08 |
| Energy contribution | -16.83 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7187720 97 + 23771897 GACGUUGGCAAAUGCCACAACUGAGGGUUUUAAGCC-CCC----------------AGUGGGUGUGGCAAAGCCCACGAAGUGCCAGCAGGAGGCCAAAUAGAAGCCAAGCAAA ...(((((((...(((...((((.((((.....)))-).)----------------))).)))((((......))))....)))))))....(((.........)))....... ( -34.90) >DroPse_CAF1 54692 80 + 1 GACGUUGGCAAAUGCCAG-----CGGC------GCCUCCCC---------------AG-GGGCGUGGAAAAGCCC-CAAAGUGUCAAGA------AAAACUGAAAUUAUGAAGA ...((((((....)))))-----)(((------((......---------------.(-((((........))))-)...)))))....------................... ( -25.40) >DroSim_CAF1 51215 98 + 1 GACGUUGGCAAAUGCCACAACUGAGGGUUUUUAGCCCCCC----------------AGUGGGCGUGGCAAAGCCCACGAAGUGCCGGCAGGAGACCAAAUAGAAGCCAAGCAAA ....(((((...((((...(((..((((.....))))...----------------.((((((........))))))..)))...))))(....).........)))))..... ( -34.20) >DroYak_CAF1 54870 97 + 1 GACGUUGGCAAAUGCCACAACUGAGGGUUUUCCGAC-CCC----------------AGUGGGCGUGGCAAAGCCCACGAAGUGCCAGCAGGCGGCCAAAUAGAAGCCAAGCAAA ...(((((((...(((...((((.(((((....)))-)))----------------))).)))((((......))))....))))))).(((..(......)..)))....... ( -39.20) >DroAna_CAF1 49317 103 + 1 GAUGUU-------GCCACAACUGCGACU----GGCCUCCUCGGGGGCCGGCGAGAGUGUGGGCGUGGUCCGGCCCCCGAAGUGCCAGCUGAAAGCCAAACAGAAGCCAAGCAAA ...(((-------((.......)))))(----(((((..(((((((((.((....))..((((...))))))))))))))).))))(((....((.........))..)))... ( -41.30) >DroPer_CAF1 63800 80 + 1 GACGUUGGCAAAUGCCAG-----CGGC------GCCUCCCC---------------AG-GGGCGUGGAAAAGCCC-CAAAGUGUCAAGA------AAAACUGAAAUUAUGAAGA ...((((((....)))))-----)(((------((......---------------.(-((((........))))-)...)))))....------................... ( -25.40) >consensus GACGUUGGCAAAUGCCACAACUGAGGCU_____GCCUCCC________________AGUGGGCGUGGCAAAGCCCACGAAGUGCCAGCAGGAGGCCAAACAGAAGCCAAGCAAA ...(((((((....((........))...............................((((((........))))))....))))))).......................... (-16.08 = -16.83 + 0.75)

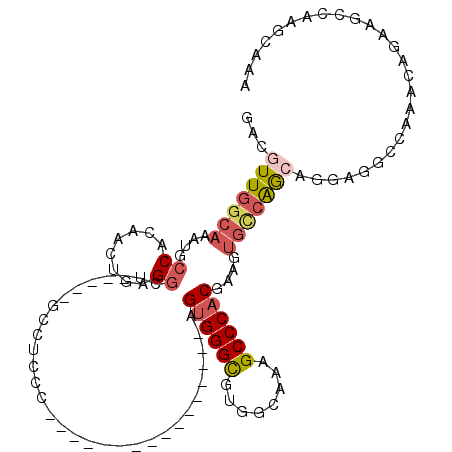
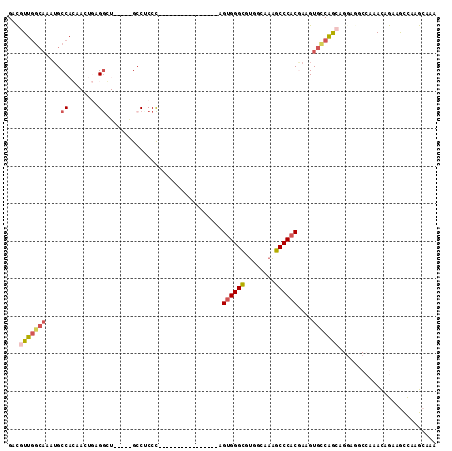
| Location | 7,187,720 – 7,187,817 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 66.91 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -12.20 |
| Energy contribution | -13.07 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.34 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7187720 97 - 23771897 UUUGCUUGGCUUCUAUUUGGCCUCCUGCUGGCACUUCGUGGGCUUUGCCACACCCACU----------------GGG-GGCUUAAAACCCUCAGUUGUGGCAUUUGCCAACGUC ..((((.((((.......)))).......))))...(((.(((..(((((((...(((----------------(((-((.......)))))))))))))))...))).))).. ( -39.00) >DroPse_CAF1 54692 80 - 1 UCUUCAUAAUUUCAGUUUU------UCUUGACACUUUG-GGGCUUUUCCACGCCC-CU---------------GGGGAGGC------GCCG-----CUGGCAUUUGCCAACGUC ...................------....(((.....(-(.((((..(((.....-.)---------------))..))))------.))(-----.((((....)))).)))) ( -24.40) >DroSim_CAF1 51215 98 - 1 UUUGCUUGGCUUCUAUUUGGUCUCCUGCCGGCACUUCGUGGGCUUUGCCACGCCCACU----------------GGGGGGCUAAAAACCCUCAGUUGUGGCAUUUGCCAACGUC ..((((.(((................)))))))...(((.(((..(((((((...(((----------------(((((........)))))))))))))))...))).))).. ( -40.09) >DroYak_CAF1 54870 97 - 1 UUUGCUUGGCUUCUAUUUGGCCGCCUGCUGGCACUUCGUGGGCUUUGCCACGCCCACU----------------GGG-GUCGGAAAACCCUCAGUUGUGGCAUUUGCCAACGUC .......((((.......))))(((....)))....(((.(((..(((((((...(((----------------(((-((......)))).)))))))))))...))).))).. ( -39.60) >DroAna_CAF1 49317 103 - 1 UUUGCUUGGCUUCUGUUUGGCUUUCAGCUGGCACUUCGGGGGCCGGACCACGCCCACACUCUCGCCGGCCCCCGAGGAGGCC----AGUCGCAGUUGUGGC-------AACAUC .((((..((((.......))))....((((((.(((((((((((((.....(......).....)))))))))))))..)))----))).))))..(((..-------..))). ( -47.80) >DroPer_CAF1 63800 80 - 1 UCUUCAUAAUUUCAGUUUU------UCUUGACACUUUG-GGGCUUUUCCACGCCC-CU---------------GGGGAGGC------GCCG-----CUGGCAUUUGCCAACGUC ...................------....(((.....(-(.((((..(((.....-.)---------------))..))))------.))(-----.((((....)))).)))) ( -24.40) >consensus UUUGCUUGGCUUCUAUUUGGCCUCCUGCUGGCACUUCGUGGGCUUUGCCACGCCCACU________________GGGAGGC_____ACCCGCAGUUGUGGCAUUUGCCAACGUC .............................(((.....((((((........))))))........................................((((....))))..))) (-12.20 = -13.07 + 0.86)
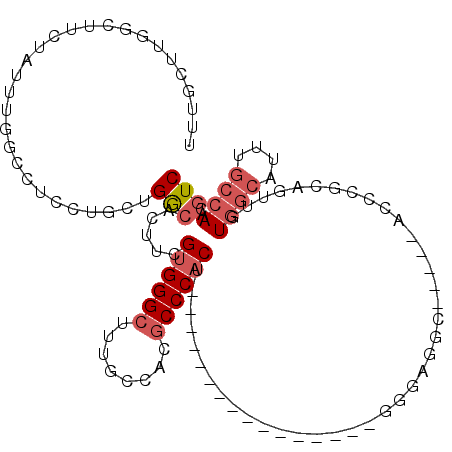
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:53 2006