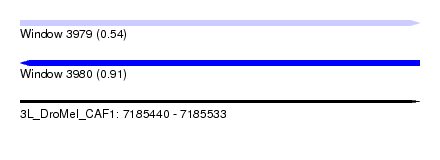
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,185,440 – 7,185,533 |
| Length | 93 |
| Max. P | 0.910984 |

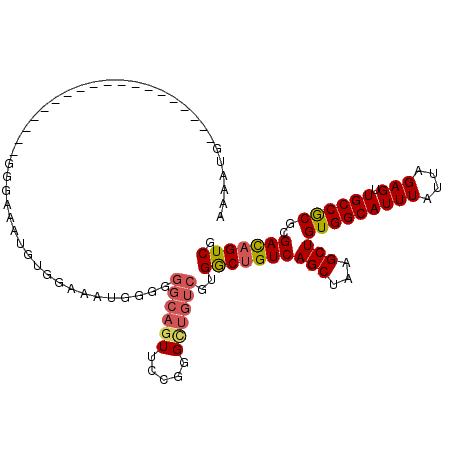
| Location | 7,185,440 – 7,185,533 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.86 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -10.52 |
| Energy contribution | -10.72 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7185440 93 + 23771897 CGACUGUCGCGCGGCAACUCUAAUAAAUGCCACAGCUUAGCUGACAGCCCCGACAGCCCGGAACUGCCCCCCAUUUCCACAUUUCCC-------------------CAUUUU ...((((((.((((((...........)))).((((...))))...))..))))))...((((.((.....)).)))).........-------------------...... ( -22.40) >DroSec_CAF1 48258 92 + 1 CGACUGUCGCGCGGCAACUCUAAUAAAUGCCACAGCUUAGCUGACAGCCACGACAGCCCGGAACUGCCC-CCAUUUCCACAUUUCCC-------------------CAUUUC ...((((((.((((((...........)))).((((...))))...))..))))))...((((.((...-.)).)))).........-------------------...... ( -22.70) >DroSim_CAF1 48862 93 + 1 CGACUGUCGCGCGGCAACUCUAAUAAAUGCCACAGCUUAGCUGACAGCCGCGACAGCCCGGAACUGCCCCCCAUUUCCACAUUUCCC-------------------CAUUUU ...(((((((((((((...........)))).((((...))))...).))))))))...((((.((.....)).)))).........-------------------...... ( -26.90) >DroEre_CAF1 56465 93 + 1 CGACUAUCGCGUGGCAGCUCUAAUAAAUGCCACAGCUUAGCUGACAGCCACGACAGCCCGCAACUGCCACCCAAUUCCCCAUCUCCC-------------------CAUUUU ........((((((((...........)))))).((...((((.(......).))))..))....))....................-------------------...... ( -17.10) >DroYak_CAF1 52388 112 + 1 CGACUAUCGCGCGGCAACUCUAAUAAAUGCCACAGCUUAGCUGACAGCCACGACAACCCGGAACUGCCACCCAUUUGCCCAUUUCCCCCCCAUUUCCACAUUUUUCCAUUUU ......(((.((((((...........)))).((((...))))...))..)))......((((.((.((......))..)).)))).......................... ( -14.10) >DroAna_CAF1 47347 78 + 1 CGACUGUCGCGUGGCAGCUCUAAUAAAUGCCACAGCUUAGCUGACGGUC--------------CUGCCCCCUAGACCCUCUAUUC-C-------------------UGCAUC .(((((((..((((((...........))))))(((...))))))))))--------------.(((....(((.....)))...-.-------------------.))).. ( -22.80) >consensus CGACUGUCGCGCGGCAACUCUAAUAAAUGCCACAGCUUAGCUGACAGCCACGACAGCCCGGAACUGCCCCCCAUUUCCACAUUUCCC___________________CAUUUU .(.(((((..(.((((...........)))).)(((...)))))))).)............................................................... (-10.52 = -10.72 + 0.20)

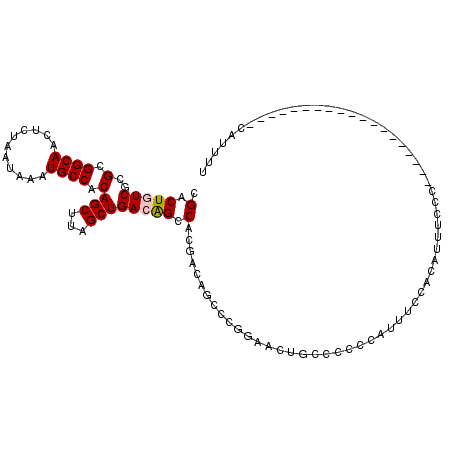
| Location | 7,185,440 – 7,185,533 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.86 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -23.87 |
| Energy contribution | -24.22 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7185440 93 - 23771897 AAAAUG-------------------GGGAAAUGUGGAAAUGGGGGGCAGUUCCGGGCUGUCGGGGCUGUCAGCUAAGCUGUGGCAUUUAUUAGAGUUGCCGCGCGACAGUCG ......-------------------.((((.(((...........))).)))).((((((((.((((........))))((((((...........)))))).)))))))). ( -31.60) >DroSec_CAF1 48258 92 - 1 GAAAUG-------------------GGGAAAUGUGGAAAUGG-GGGCAGUUCCGGGCUGUCGUGGCUGUCAGCUAAGCUGUGGCAUUUAUUAGAGUUGCCGCGCGACAGUCG ......-------------------.((((.(((........-..))).)))).(((((((((((((........))))((((((...........))))))))))))))). ( -33.00) >DroSim_CAF1 48862 93 - 1 AAAAUG-------------------GGGAAAUGUGGAAAUGGGGGGCAGUUCCGGGCUGUCGCGGCUGUCAGCUAAGCUGUGGCAUUUAUUAGAGUUGCCGCGCGACAGUCG ......-------------------.((((.(((...........))).)))).(((((((((((((........))))((((((...........))))))))))))))). ( -35.40) >DroEre_CAF1 56465 93 - 1 AAAAUG-------------------GGGAGAUGGGGAAUUGGGUGGCAGUUGCGGGCUGUCGUGGCUGUCAGCUAAGCUGUGGCAUUUAUUAGAGCUGCCACGCGAUAGUCG ......-------------------....(((......((((.(((((((..(((....)))..))))))).))))((.(((((((((....))).))))))))....))). ( -33.90) >DroYak_CAF1 52388 112 - 1 AAAAUGGAAAAAUGUGGAAAUGGGGGGGAAAUGGGCAAAUGGGUGGCAGUUCCGGGUUGUCGUGGCUGUCAGCUAAGCUGUGGCAUUUAUUAGAGUUGCCGCGCGAUAGUCG .................................(((...((.((((((..((..(..(((((..(((........)))..)))))....)..))..)))))).))...))). ( -31.50) >DroAna_CAF1 47347 78 - 1 GAUGCA-------------------G-GAAUAGAGGGUCUAGGGGGCAG--------------GACCGUCAGCUAAGCUGUGGCAUUUAUUAGAGCUGCCACGCGACAGUCG (((.(.-------------------.-.....((.(((((........)--------------)))).))......((.(((((((((....))).)))))))))...))). ( -23.30) >consensus AAAAUG___________________GGGAAAUGUGGAAAUGGGGGGCAGUUCCGGGCUGUCGUGGCUGUCAGCUAAGCUGUGGCAUUUAUUAGAGUUGCCGCGCGACAGUCG ............................................((((((.....))))))..((((((((((...)))(((((((((....))).))))))..))))))). (-23.87 = -24.22 + 0.35)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:47 2006