
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,169,724 – 7,169,814 |
| Length | 90 |
| Max. P | 0.711515 |

| Location | 7,169,724 – 7,169,814 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.85 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -12.52 |
| Energy contribution | -12.52 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
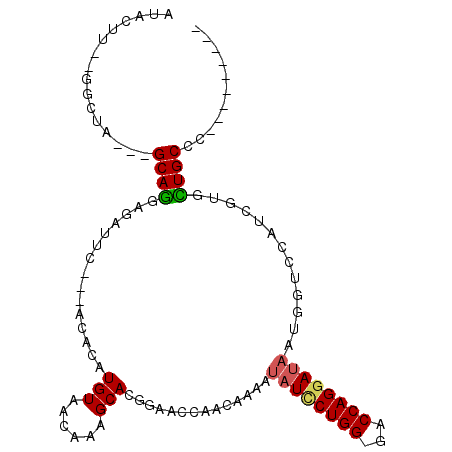
>3L_DroMel_CAF1 7169724 90 + 23771897 AUACUU--GGCUC---GCAGGAGAUUC---ACACAUGUAACAAAGCACGGAACCAACAAAAUAUCCUGG-GACCAGAAUAAUGGUCCAUCGUGCUGCCC--------- ......--(((..---(((........---.....))).....(((((((......((........))(-(((((......)))))).)))))))))).--------- ( -23.22) >DroSec_CAF1 32380 90 + 1 AUACUU--GGCUA---GCAGGAGAUUC---ACACAUGUAACAAAGCACGGAACCAACAAAAUAUCCUGG-GACCAGGAUAAUGGUCCAGCGUGCUGCCC--------- ......--(((.(---(((.(......---.....(((......))).(((.(((......(((((((.-...))))))).))))))..).))))))).--------- ( -26.10) >DroSim_CAF1 32773 90 + 1 AUACUU--GGCUA---GCAGGAGAUUC---ACACAUGUAACAAAGCACGGAACCAACAAAAUAUCCUGG-GACCAGGAUAAUGGUCCAUCGUGCUGCCC--------- ......--(((.(---(((.((.....---.....(((......))).(((.(((......(((((((.-...))))))).)))))).)).))))))).--------- ( -27.80) >DroEre_CAF1 37298 90 + 1 AUACUU--GGGUA---GCAGCAGAUUC---ACACAUGUAACAAAGCAUCGAACGAACAAAAUAUCCUGG-GACCAGGAUAAUGCUCCAUCGUGUUGCCC--------- ......--(((((---((((((.....---.....))).....((((((....))......(((((((.-...))))))).))))......))))))))--------- ( -23.80) >DroYak_CAF1 35547 102 + 1 AUACUU--GGGUA---GCAGCAGAUUCUGCUCACAUGUAACAAAGCAUCGAAAGAACAAAAUAUCCUGG-GACCAGGAUAAUGCUCCAUCGUGUUGCCCCGUUUUUUU ......--(((((---(((((((...)))).............((((((....))......(((((((.-...))))))).))))......))))))))......... ( -31.40) >DroPer_CAF1 42872 81 + 1 GUACUCCCAAGAAGGGGCAAGAUUUUC---ACACAUGUAACAAAGCAU---------------UUCUGGCAGCCAGGAUAAUGGGCCAUCGGCUUGCCU--------- ......((.....))((((((......---....((((......))))---------------...((((..(((......)))))))....)))))).--------- ( -18.50) >consensus AUACUU__GGCUA___GCAGGAGAUUC___ACACAUGUAACAAAGCACGGAACCAACAAAAUAUCCUGG_GACCAGGAUAAUGGUCCAUCGUGCUGCCC_________ ................((((...............(((......)))..............((((((((...)))))))).............))))........... (-12.52 = -12.52 + 0.00)


| Location | 7,169,724 – 7,169,814 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.85 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -11.14 |
| Energy contribution | -11.39 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.38 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
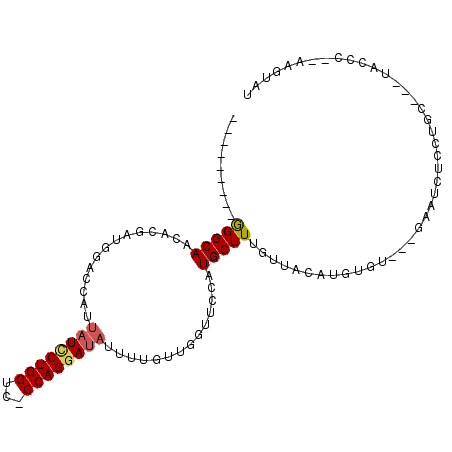
>3L_DroMel_CAF1 7169724 90 - 23771897 ---------GGGCAGCACGAUGGACCAUUAUUCUGGUC-CCAGGAUAUUUUGUUGGUUCCGUGCUUUGUUACAUGUGU---GAAUCUCCUGC---GAGCC--AAGUAU ---------.(((((((((..((((((.(((((((...-.)))))))......))))))))))))...((((....))---)).........---..)))--...... ( -28.60) >DroSec_CAF1 32380 90 - 1 ---------GGGCAGCACGCUGGACCAUUAUCCUGGUC-CCAGGAUAUUUUGUUGGUUCCGUGCUUUGUUACAUGUGU---GAAUCUCCUGC---UAGCC--AAGUAU ---------.(((((((((..((((((.(((((((...-.)))))))......))))))))))))...((((....))---)).........---..)))--...... ( -31.50) >DroSim_CAF1 32773 90 - 1 ---------GGGCAGCACGAUGGACCAUUAUCCUGGUC-CCAGGAUAUUUUGUUGGUUCCGUGCUUUGUUACAUGUGU---GAAUCUCCUGC---UAGCC--AAGUAU ---------.(((((((((..((((((.(((((((...-.)))))))......))))))))))))...((((....))---)).........---..)))--...... ( -31.50) >DroEre_CAF1 37298 90 - 1 ---------GGGCAACACGAUGGAGCAUUAUCCUGGUC-CCAGGAUAUUUUGUUCGUUCGAUGCUUUGUUACAUGUGU---GAAUCUGCUGC---UACCC--AAGUAU ---------(((.....(((..(((((.(((((((...-.)))))))...)))))..)))..((....((((....))---))....))...---..)))--...... ( -27.90) >DroYak_CAF1 35547 102 - 1 AAAAAAACGGGGCAACACGAUGGAGCAUUAUCCUGGUC-CCAGGAUAUUUUGUUCUUUCGAUGCUUUGUUACAUGUGAGCAGAAUCUGCUGC---UACCC--AAGUAU .....(((((((((...(((.((((((.(((((((...-.)))))))...)))))).))).)))))))))((.((.((((((......))))---).).)--).)).. ( -37.20) >DroPer_CAF1 42872 81 - 1 ---------AGGCAAGCCGAUGGCCCAUUAUCCUGGCUGCCAGAA---------------AUGCUUUGUUACAUGUGU---GAAAAUCUUGCCCCUUCUUGGGAGUAC ---------.((((.((((((((....)))))..)))))))....---------------........((((....))---))......((((((.....))).))). ( -19.80) >consensus _________GGGCAACACGAUGGACCAUUAUCCUGGUC_CCAGGAUAUUUUGUUGGUUCCAUGCUUUGUUACAUGUGU___GAAUCUCCUGC___UACCC__AAGUAU .........(((((..............((((((((...))))))))..............))))).......................................... (-11.14 = -11.39 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:41 2006