| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,151,026 – 7,151,128 |
| Length | 102 |
| Max. P | 0.951426 |

| Location | 7,151,026 – 7,151,128 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 86.35 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -22.77 |
| Energy contribution | -23.90 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7151026 102 + 23771897 UGCCACUUGCAUCUUGGCCCCUGCCACACU------UUCAAGUUGCAGAAGCGAAAAGUUUCGUGGCAAAAGUUUAUGAAAAAUUAUGAAUUUCAAUUUAGCCUAAUC ((((((.((((.((((((....))))....------....)).))))(((((.....))))))))))).((((((((((....))))))))))............... ( -26.80) >DroSec_CAF1 13708 102 + 1 UGCCACCUGCAGCUUGGCCCCGGCCACAUU------UUCAAGUGGCAGAAGCGAAAAGUUUCGUGGCAAAAGUUUAUGAAAAAUUAUGAAUUUCAAUUUAGCCGAAUC ((((((.((.....((((....))))....------..)).))))))(((((.....))))).((((((((((((((((....)))))))))....))).)))).... ( -29.00) >DroEre_CAF1 18417 102 + 1 UGCCACUUGCAGCUUGGCCCCUUCCACUGC------GCCAAGUGGCAGAGGCGAAAAGUUUCGUGGCAAAAGCUUAUGAGAAAUUAUGAAUUUCAAUUUAGCCGAAUC ((((((.(((.(((((((.(........).------))))))).)))(((((.....)))))))))))...(((.....(((((.....))))).....)))...... ( -32.70) >DroYak_CAF1 16053 108 + 1 UGCCACUUGCAGCUUGGCCCUCUCCACUCUUUCUCUCUCAAGUGGCAAAAGCGAAAAGUUUCGUGGCUUAAGCUUAUGAAAUAUUAUGAAUUUCAAUUUAGCCAAAUC (((((((((.((..(((......)))..))........)))))))))...(((((....)))))((((.((.....((((((.......)))))).)).))))..... ( -24.10) >consensus UGCCACUUGCAGCUUGGCCCCUGCCACACU______UUCAAGUGGCAGAAGCGAAAAGUUUCGUGGCAAAAGCUUAUGAAAAAUUAUGAAUUUCAAUUUAGCCGAAUC .(((((.(((.((((((....................)))))).)))(((((.....))))))))))..((((((((((....))))))))))............... (-22.77 = -23.90 + 1.13)

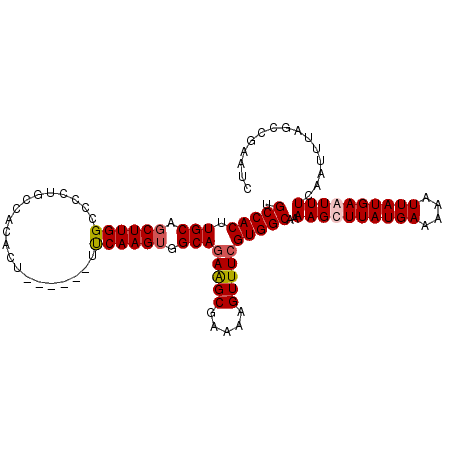

| Location | 7,151,026 – 7,151,128 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 86.35 |
| Mean single sequence MFE | -29.95 |
| Consensus MFE | -24.24 |
| Energy contribution | -25.93 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7151026 102 - 23771897 GAUUAGGCUAAAUUGAAAUUCAUAAUUUUUCAUAAACUUUUGCCACGAAACUUUUCGCUUCUGCAACUUGAA------AGUGUGGCAGGGGCCAAGAUGCAAGUGGCA .............(((((.........)))))........((((((.....((((.((((((((.(((....------)))...)))))))).)))).....)))))) ( -26.60) >DroSec_CAF1 13708 102 - 1 GAUUCGGCUAAAUUGAAAUUCAUAAUUUUUCAUAAACUUUUGCCACGAAACUUUUCGCUUCUGCCACUUGAA------AAUGUGGCCGGGGCCAAGCUGCAGGUGGCA .....(((.(((.(((((.........))))).....))).))).((((....)))).....((((((((..------....((((....)))).....)))))))). ( -29.00) >DroEre_CAF1 18417 102 - 1 GAUUCGGCUAAAUUGAAAUUCAUAAUUUCUCAUAAGCUUUUGCCACGAAACUUUUCGCCUCUGCCACUUGGC------GCAGUGGAAGGGGCCAAGCUGCAAGUGGCA .....((((.....(((((.....))))).....))))..(((((((((....)))((((((.(((((....------..))))).))))))..........)))))) ( -31.90) >DroYak_CAF1 16053 108 - 1 GAUUUGGCUAAAUUGAAAUUCAUAAUAUUUCAUAAGCUUAAGCCACGAAACUUUUCGCUUUUGCCACUUGAGAGAGAAAGAGUGGAGAGGGCCAAGCUGCAAGUGGCA .....((((....((((((.......))))))..))))...(((((....((((((((((((....(((....))).)))))))))))).((......))..))))). ( -32.30) >consensus GAUUCGGCUAAAUUGAAAUUCAUAAUUUUUCAUAAACUUUUGCCACGAAACUUUUCGCUUCUGCCACUUGAA______AGAGUGGAAGGGGCCAAGCUGCAAGUGGCA .....((((....(((((.........)))))..))))...((((((((....)))(((((((((((((..........)))))))))))))..........))))). (-24.24 = -25.93 + 1.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:32 2006