| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,130,353 – 7,130,450 |
| Length | 97 |
| Max. P | 0.762890 |

| Location | 7,130,353 – 7,130,450 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 85.28 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -19.56 |
| Energy contribution | -20.12 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7130353 97 + 23771897 GCAGCUGGAGAAGAAGAAGAGCAUGAAUGAUCGCUCAAGGACUCGGUAUCGUCCUGCCGUUUCCUCUCUCU------CUUUCUCUCUCUGGUUCACUUCCUUU (.(((..((((.((..(((((...((.(((....))).(((..(((((......)))))..)))))...))------)))..))))))..))))......... ( -30.00) >DroSec_CAF1 45195 91 + 1 GCAGCUGGAGAAGAAGAAGAGCAUAAAUGAUCGCUCAAGGACUCGAAAUCGUCCUGCCCUUUCCUCUCUC------------UCUCUCUGGUUCACUUCCUUU (.(((..((((((((((.((((..........)))).(((((........))))).........))).))------------).))))..))))......... ( -24.60) >DroEre_CAF1 45475 95 + 1 GCAGCUGGAGAAGAAGAAGAGCACGAAUGAUCGCUCAAGGACUCGGUAUCGUCCUGCUGUUUCCUCUCUC--------UCUCCCUGUCUUGCUCACUUGCUUU ((((..((((((((.(((((((.(((....)))....(((((........)))))))).)))).))).))--------)))..))))...((......))... ( -26.10) >DroYak_CAF1 47290 103 + 1 GCAGCUGGAGAAGAAGAAGAGCGCGAAUGAUCGCUCAAGGACUCGAAAUCGUCCUGCUGUUUCCUCUCUCGCUCUCGCUCUCUCUCUCUUGUUCAGUGCCUUU (((.(((((.((((((((((((((((..((..((...(((((........)))))...))....))..))))....))))).))).)))).)))))))).... ( -35.20) >consensus GCAGCUGGAGAAGAAGAAGAGCACGAAUGAUCGCUCAAGGACUCGAAAUCGUCCUGCCGUUUCCUCUCUC________UCUCUCUCUCUGGUUCACUUCCUUU (.((((((((((((.(((((((..........)))).(((((........))))).....))).))).................))))))))))......... (-19.56 = -20.12 + 0.56)

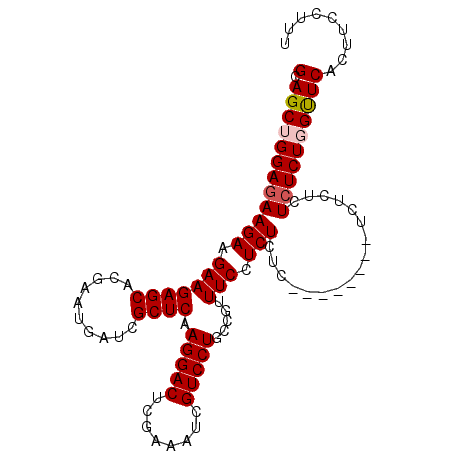

| Location | 7,130,353 – 7,130,450 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 85.28 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -22.46 |
| Energy contribution | -22.28 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7130353 97 - 23771897 AAAGGAAGUGAACCAGAGAGAGAAAG------AGAGAGAGGAAACGGCAGGACGAUACCGAGUCCUUGAGCGAUCAUUCAUGCUCUUCUUCUUCUCCAGCUGC .............(((...(((((..------((((.(((((..(((..........)))..)))))(((((........)))))))))..)))))...))). ( -26.60) >DroSec_CAF1 45195 91 - 1 AAAGGAAGUGAACCAGAGAGA------------GAGAGAGGAAAGGGCAGGACGAUUUCGAGUCCUUGAGCGAUCAUUUAUGCUCUUCUUCUUCUCCAGCUGC .............(((...((------------((.(((((((.(((((((((........)))).(((....)))....))))))))))))))))...))). ( -28.10) >DroEre_CAF1 45475 95 - 1 AAAGCAAGUGAGCAAGACAGGGAGA--------GAGAGAGGAAACAGCAGGACGAUACCGAGUCCUUGAGCGAUCAUUCGUGCUCUUCUUCUUCUCCAGCUGC ..(((..((.......))..(((((--------(..((((((....(((((((........)))))...((((....))))))))))))..)))))).))).. ( -30.70) >DroYak_CAF1 47290 103 - 1 AAAGGCACUGAACAAGAGAGAGAGAGCGAGAGCGAGAGAGGAAACAGCAGGACGAUUUCGAGUCCUUGAGCGAUCAUUCGCGCUCUUCUUCUUCUCCAGCUGC ....((((((.....(((((..((((.(((.(((((.(((....).(((((((........)))))...))..)).))))).)))))))..)))))))).))) ( -35.80) >consensus AAAGGAAGUGAACAAGAGAGAGAGA________GAGAGAGGAAACAGCAGGACGAUACCGAGUCCUUGAGCGAUCAUUCAUGCUCUUCUUCUUCUCCAGCUGC .................................((((((((((.....(((((........))))).(((((........))))))))))).))))....... (-22.46 = -22.28 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:21 2006