
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,126,682 – 7,126,779 |
| Length | 97 |
| Max. P | 0.999126 |

| Location | 7,126,682 – 7,126,779 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 95.05 |
| Mean single sequence MFE | -29.89 |
| Consensus MFE | -26.49 |
| Energy contribution | -26.33 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.999126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
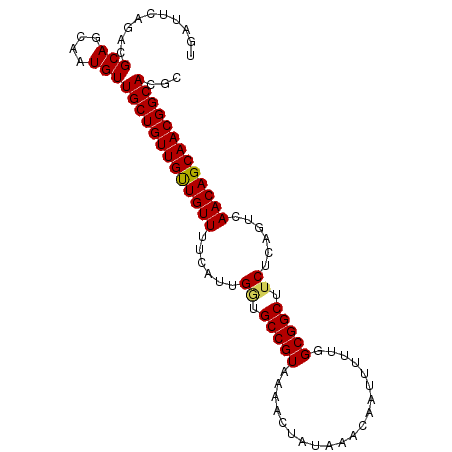
>3L_DroMel_CAF1 7126682 97 + 23771897 UGAUUCAGACGCAGCAAUGUUGCUGUUGUUGUUUUCUUUGGUGCCGUAAAACUAUAAACAAUUUUUGGCGGCUUCUCAGUCAACAGCAACGGCACGC ..........(((....)))(((((((((((((..((..((.(((((.(((............))).))))).))..))..)))))))))))))... ( -29.60) >DroSec_CAF1 41434 97 + 1 UGAUUCAGACGCAGCAAUGUUGCUGUUGUUGUUUUCAUUGGUGCCGUAAAACUAUAAACAAUUUUUGGCGGCUUCUCAGUCAACAGCAACGGCACGC ..........(((....)))(((((((((((((...(((((.(((((.(((............))).)))))...))))).)))))))))))))... ( -28.20) >DroSim_CAF1 42180 97 + 1 UGAUUCAGACGCAGCAAUGUUGCUGUUGUUGUUUUCAUUGGUGCCGUAAAACUAUAAACCAUUUUUGGCGGCUUCUCAGUCAACAGCAACGGCACGC (((....(((((((((....))))).))))....)))...(((((((...........(((....))).((((....)))).......))))))).. ( -28.30) >DroEre_CAF1 41564 96 + 1 UGAUUCAGACGCAGCAAUGUUGCUGUUGCUGUUUUCGUUGCUGCCGUAA-ACUAUAAACAAUUUUUGGCGGCUUCUCAGUCAACAGCAACGGCACGC ..........(((....)))(((((((((((((..(...(((((((...-...............)))))))......)..)))))))))))))... ( -32.37) >DroYak_CAF1 43311 91 + 1 UGAUUCAGACGCAGCAAUGUUGCUGUUGCUGUU-----UGAUGCCGUAA-ACUAUAAACAAUUUUUGGCGGCUUCUCAGUCAACAGCAACGGCACGC .............((...(((((((((((((..-----.((.(((((..-...((.....)).....))))).)).))).)))))))))).)).... ( -31.00) >consensus UGAUUCAGACGCAGCAAUGUUGCUGUUGUUGUUUUCAUUGGUGCCGUAAAACUAUAAACAAUUUUUGGCGGCUUCUCAGUCAACAGCAACGGCACGC ....................(((((((((((((......((.(((((....................))))).))......)))))))))))))... (-26.49 = -26.33 + -0.16)

| Location | 7,126,682 – 7,126,779 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 95.05 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -23.64 |
| Energy contribution | -24.24 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7126682 97 - 23771897 GCGUGCCGUUGCUGUUGACUGAGAAGCCGCCAAAAAUUGUUUAUAGUUUUACGGCACCAAAGAAAACAACAACAGCAACAUUGCUGCGUCUGAAUCA (((.((.((((((((((........((((...(((((((....))))))).))))......(....)..))))))))))...))))).......... ( -25.90) >DroSec_CAF1 41434 97 - 1 GCGUGCCGUUGCUGUUGACUGAGAAGCCGCCAAAAAUUGUUUAUAGUUUUACGGCACCAAUGAAAACAACAACAGCAACAUUGCUGCGUCUGAAUCA (((.((.((((((((((........((((...(((((((....))))))).)))).....((....)).))))))))))...))))).......... ( -26.70) >DroSim_CAF1 42180 97 - 1 GCGUGCCGUUGCUGUUGACUGAGAAGCCGCCAAAAAUGGUUUAUAGUUUUACGGCACCAAUGAAAACAACAACAGCAACAUUGCUGCGUCUGAAUCA (((.((.((((((((((......((((((.......))))))...(((((.((.......)))))))..))))))))))...))))).......... ( -25.80) >DroEre_CAF1 41564 96 - 1 GCGUGCCGUUGCUGUUGACUGAGAAGCCGCCAAAAAUUGUUUAUAGU-UUACGGCAGCAACGAAAACAGCAACAGCAACAUUGCUGCGUCUGAAUCA (((.((.((((((((((.(((.(..((.(((..((((((....))))-))..))).))..).....)))))))))))))...))))).......... ( -30.70) >DroYak_CAF1 43311 91 - 1 GCGUGCCGUUGCUGUUGACUGAGAAGCCGCCAAAAAUUGUUUAUAGU-UUACGGCAUCA-----AACAGCAACAGCAACAUUGCUGCGUCUGAAUCA (((.((.((((((((((.(((.((.((((....((((((....))))-)).)))).)).-----..)))))))))))))...))))).......... ( -30.50) >consensus GCGUGCCGUUGCUGUUGACUGAGAAGCCGCCAAAAAUUGUUUAUAGUUUUACGGCACCAAUGAAAACAACAACAGCAACAUUGCUGCGUCUGAAUCA (((.((.((((((((((.((....))..(((.(((((((....)))))))..)))..............))))))))))...))))).......... (-23.64 = -24.24 + 0.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:17 2006