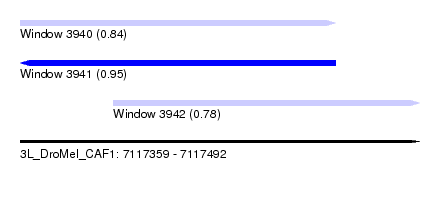
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,117,359 – 7,117,492 |
| Length | 133 |
| Max. P | 0.948359 |

| Location | 7,117,359 – 7,117,464 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 73.01 |
| Mean single sequence MFE | -26.06 |
| Consensus MFE | -12.18 |
| Energy contribution | -12.57 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7117359 105 + 23771897 AAUACUUGAAUCCGAAUACUUGGUAGCCCAUUACUGCAGGGCUUGAGAAAUAGAUU--UUCAGACGAAAAGUAUGCAGCAUU-GAAA-GUCUAAAUGCUGCAGGGUAUG- .........((((..((((((.((.((((.........))))(((((((.....))--)))))))...))))))((((((((-....-.....)))))))).))))...- ( -29.30) >DroVir_CAF1 63314 85 + 1 AAUACUU---UUCAAAUGCAUGGCAC----UAAAUGCAGCCU--G---------------UUGCCCAAAUGUAUGCAACACAAAUC-UGACAGCGCGCUCGAAAGUAUGC .((((((---((....(((((.....----...)))))((((--(---------------((.......(((.....)))......-.))))).))....)))))))).. ( -18.14) >DroSec_CAF1 32337 105 + 1 AAUACUUGUAUCCGAAUACUUGGUAGCCCAUUACUGCAUGGCUUGAGAAAUAGAUU--UUCAGACGAAAAGUAUGCAGCAUU-GAAA-GCCUAAAUGCUGCAGGGUAUG- ..((((.((((....))))..))))((((..((((...((..(((((((.....))--))))).))...))))(((((((((-....-.....)))))))))))))...- ( -29.30) >DroSim_CAF1 33071 105 + 1 AAUACUUGUAUCCGAAUACUUGGUAGCCCAUUACUGCAGGCCUUGAGAAAUAGAUU--UUCAGACGAAAAGUAUGCAGCAUU-GAAA-GUCUAAAUGCUGCAGGGUAUG- .......((((((..(((((((((.((........))..)))(((((((.....))--))))).....))))))((((((((-....-.....)))))))).)))))).- ( -28.60) >DroEre_CAF1 32364 98 + 1 AAUACUUGAAUCCGAAUACUUGGUAACCCAG-------GGGCUUGAGAACUAUAUU--UUCAGAUGAAAAGUAUGCAGCAUU-GAAA-GUCCAAAUGCUGCAGGGUAUG- .((((((..(((......(((((....))))-------)....((((((.....))--)))))))...))))))((((((((-....-.....))))))))........- ( -25.90) >DroYak_CAF1 33973 100 + 1 AAUACUUGGAGCCGAAUAUUUGGUAUCCCAU-------GGGCUAGAGAAAUAUAUUUUUUUAGACGAAAAGUAUGCAGCACU-GCAA-GUCGAAAUGCUGCAGGGUAUG- .((((((((((((((....))))).)))..(-------(..(((((((((....))))))))).)).......(((((((..-.(..-...)...))))))))))))).- ( -25.10) >consensus AAUACUUGAAUCCGAAUACUUGGUAGCCCAUUACUGCAGGGCUUGAGAAAUAGAUU__UUCAGACGAAAAGUAUGCAGCAUU_GAAA_GUCUAAAUGCUGCAGGGUAUG_ .((((((....((((....))))..................................................(((((((((...........))))))))))))))).. (-12.18 = -12.57 + 0.39)

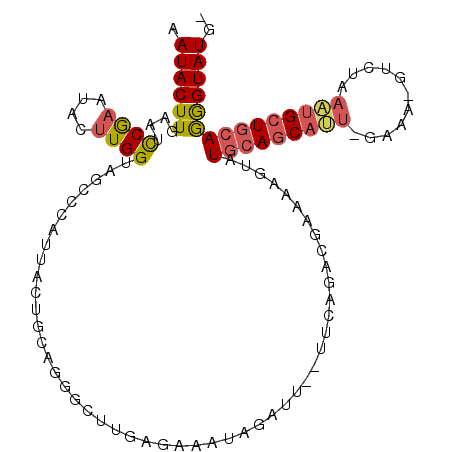
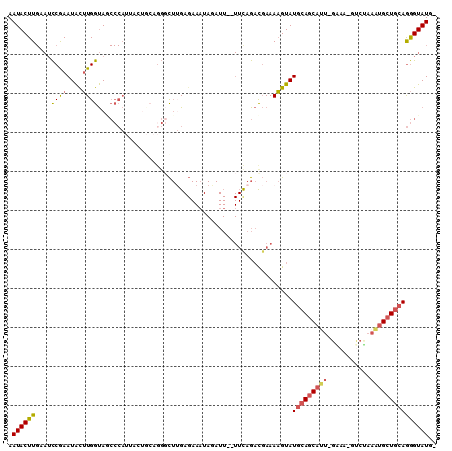
| Location | 7,117,359 – 7,117,464 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 73.01 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -12.39 |
| Energy contribution | -13.44 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7117359 105 - 23771897 -CAUACCCUGCAGCAUUUAGAC-UUUC-AAUGCUGCAUACUUUUCGUCUGAA--AAUCUAUUUCUCAAGCCCUGCAGUAAUGGGCUACCAAGUAUUCGGAUUCAAGUAUU -.......(((((((((.....-....-)))))))))(((((...(((((((--....(((((....(((((.........)))))...))))))))))))..))))).. ( -28.10) >DroVir_CAF1 63314 85 - 1 GCAUACUUUCGAGCGCGCUGUCA-GAUUUGUGUUGCAUACAUUUGGGCAA---------------C--AGGCUGCAUUUA----GUGCCAUGCAUUUGAA---AAGUAUU ..(((((((((((.(((......-......((((((...(....).))))---------------)--)(((.((.....----))))).))).)))).)---)))))). ( -21.20) >DroSec_CAF1 32337 105 - 1 -CAUACCCUGCAGCAUUUAGGC-UUUC-AAUGCUGCAUACUUUUCGUCUGAA--AAUCUAUUUCUCAAGCCAUGCAGUAAUGGGCUACCAAGUAUUCGGAUACAAGUAUU -.......(((((((((.....-....-)))))))))(((((...(((((((--....(((((....((((...((....))))))...))))))))))))..))))).. ( -25.10) >DroSim_CAF1 33071 105 - 1 -CAUACCCUGCAGCAUUUAGAC-UUUC-AAUGCUGCAUACUUUUCGUCUGAA--AAUCUAUUUCUCAAGGCCUGCAGUAAUGGGCUACCAAGUAUUCGGAUACAAGUAUU -.......(((((((((.....-....-)))))))))(((((...(((((((--....(((((.....(((((........)))))...))))))))))))..))))).. ( -25.80) >DroEre_CAF1 32364 98 - 1 -CAUACCCUGCAGCAUUUGGAC-UUUC-AAUGCUGCAUACUUUUCAUCUGAA--AAUAUAGUUCUCAAGCCC-------CUGGGUUACCAAGUAUUCGGAUUCAAGUAUU -.......(((((((((.((..-..))-)))))))))(((((...(((((((--.((...((.((((.....-------.))))..))...)).)))))))..))))).. ( -23.40) >DroYak_CAF1 33973 100 - 1 -CAUACCCUGCAGCAUUUCGAC-UUGC-AGUGCUGCAUACUUUUCGUCUAAAAAAAUAUAUUUCUCUAGCCC-------AUGGGAUACCAAAUAUUCGGCUCCAAGUAUU -.......(((((((((.(...-..).-)))))))))....................((((((....((((.-------.(((....))).......))))..)))))). ( -17.90) >consensus _CAUACCCUGCAGCAUUUAGAC_UUUC_AAUGCUGCAUACUUUUCGUCUGAA__AAUCUAUUUCUCAAGCCCUGCAGUAAUGGGCUACCAAGUAUUCGGAUACAAGUAUU ........(((((((((...........)))))))))(((((...(((((((...........((((.............))))..........)))))))..))))).. (-12.39 = -13.44 + 1.05)
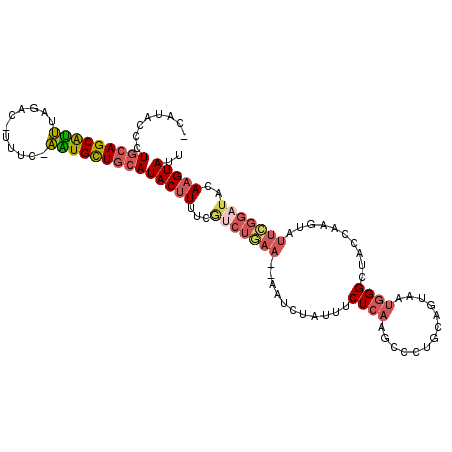
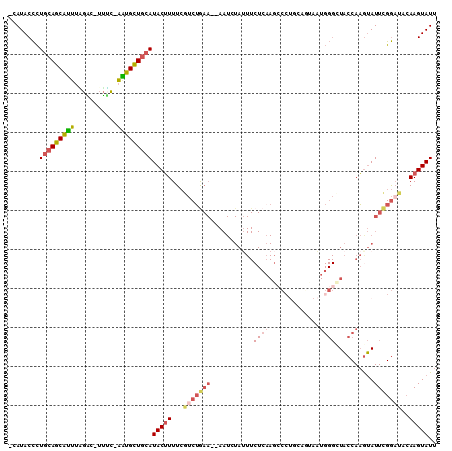
| Location | 7,117,390 – 7,117,492 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 89.91 |
| Mean single sequence MFE | -23.88 |
| Consensus MFE | -17.98 |
| Energy contribution | -18.18 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7117390 102 + 23771897 UACUGCAGGGCUUGAGAAAUAGAUU--UUCAGACGAAAAGUAUGCAGCAUUGAAAGUCUAAAUGCUGCAGGGUAUGCUAUAUUAGCUAAAUGCAAAACCAACUC ...((((.((((.((...((((.((--(((....)))))...(((((((((.........))))))))).......)))).))))))...)))).......... ( -24.80) >DroSec_CAF1 32368 102 + 1 UACUGCAUGGCUUGAGAAAUAGAUU--UUCAGACGAAAAGUAUGCAGCAUUGAAAGCCUAAAUGCUGCAGGGUAUGCUAUAUUAGCUAAAUGCAAAACCAACUC ....(((((.((((((((.....))--))))...........(((((((((.........))))))))))).))))).......((.....))........... ( -24.20) >DroSim_CAF1 33102 102 + 1 UACUGCAGGCCUUGAGAAAUAGAUU--UUCAGACGAAAAGUAUGCAGCAUUGAAAGUCUAAAUGCUGCAGGGUAUGCUAUAUUAGCUAAAUGCAAAACCAACUC ....(((.((((((((((.....))--))))...........(((((((((.........))))))))))))).))).......((.....))........... ( -25.20) >DroEre_CAF1 32395 95 + 1 -------GGGCUUGAGAACUAUAUU--UUCAGAUGAAAAGUAUGCAGCAUUGAAAGUCCAAAUGCUGCAGGGUAUGCUAUACUAGCUAAAUGCAAAACCAACUC -------((..(((...(((...((--(((....)))))...(((((((((.........))))))))).)))..((((...))))......)))..))..... ( -22.00) >DroYak_CAF1 34004 97 + 1 -------GGGCUAGAGAAAUAUAUUUUUUUAGACGAAAAGUAUGCAGCACUGCAAGUCGAAAUGCUGCAGGGUAUGCUAUAUUAGCUAAAUGCAAAACCAACUC -------((.(((((((((....)))))))))......(((((((....(((((.((......))))))).)))))))......((.....))....))..... ( -23.20) >consensus UACUGCAGGGCUUGAGAAAUAGAUU__UUCAGACGAAAAGUAUGCAGCAUUGAAAGUCUAAAUGCUGCAGGGUAUGCUAUAUUAGCUAAAUGCAAAACCAACUC .............(((..........................(((((((((.........)))))))))((...(((..............)))...))..))) (-17.98 = -18.18 + 0.20)

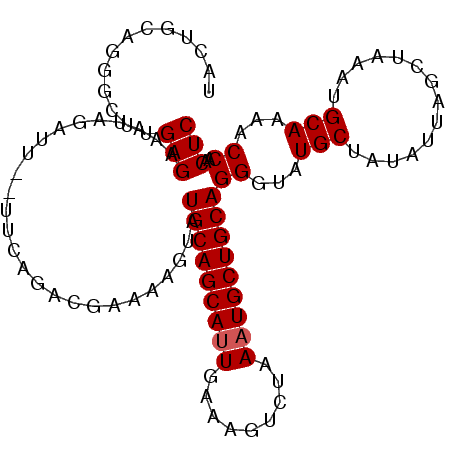
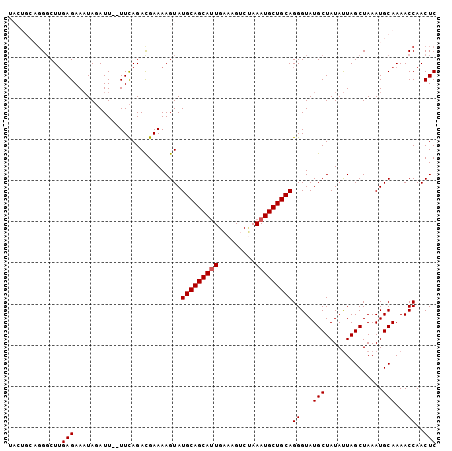
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:10 2006