
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,091,335 – 7,091,519 |
| Length | 184 |
| Max. P | 0.997709 |

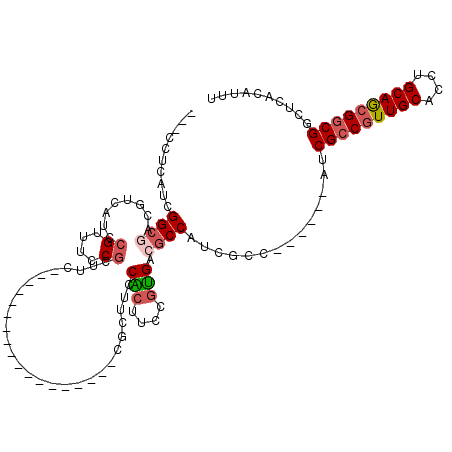
| Location | 7,091,335 – 7,091,427 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.79 |
| Mean single sequence MFE | -33.63 |
| Consensus MFE | -19.56 |
| Energy contribution | -20.20 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7091335 92 + 23771897 ---CCUCAUCGGCGACGUCACCCGUUUCCGGUUC------------------CGCUUCCACUUCCGUGACGCCAUCGCC------AUCGCCGUUGCACCUGCAGCGGCGGCUCACAUUU ---.......((((((((((((((....)))...------------------.(......)....))))))...)))))------.((((((((((....))))))))))......... ( -34.90) >DroSec_CAF1 5979 89 + 1 ---CCUCAUCGGC---GUCAUCCGUUUCCGGUUU------------------CGCUUCCACUUCCGUGACGCCAUCGCC------AUCGCCGUUGCACCUGCAGCGGCGCCUCACAUUU ---.......(((---((((((((....)))...------------------.............))))))))......------..(((((((((....))))))))).......... ( -31.19) >DroSim_CAF1 6085 89 + 1 ---CCUCAUCGGC---GUCAUCCGUUUCCGGUUU------------------CGCUUCCACUUCCGUGACGCCAUCGCC------AUCGCCGUUGCACCUGCAGCGGCGGCUCACAUUU ---.......(((---((((((((....)))...------------------.............))))))))...(((------...((((((((....)))))))))))........ ( -33.29) >DroEre_CAF1 6468 89 + 1 CCGCCACAUCGGCGACGACAUCCGUUUCCGCUUC------------------------CGCCUCCGUGACGCCGUCGCC------CUCGCCGUUGCACCUGCAGCGGCGGCUCACAUUU ..(((.....(((((((.(.((((.....((...------------------------.))...)).)).).)))))))------...((((((((....)))))))))))........ ( -37.40) >DroYak_CAF1 6823 113 + 1 UCGCCUCAUCGGCGACGUCAUCCGUUUCCGCUUCCGGUUCCGCGUCCGUUUCCGCAUCCGCUUCCGUGACGCCAUCGCC------AUCGCCGUUGCACCUGCAGCGCCGUGUCACAUUU ..(((.....)))(((((...(((..........)))....))))).......((....))....(((((((....((.------...))((((((....))))))..))))))).... ( -32.40) >DroAna_CAF1 5763 95 + 1 CGUCGUCAACGGAGGAGUCUACGGUUUCCGCUUC------------------------CGUGUCGCCGACGCCAUCGCCUGCCGCCUCGCCAUUGCACCUGCAACGGCGGCUGACAUUC ....(((((((((((((.(....).))))...))------------------------)))(((((((..((....((.....))...))..((((....))))))))))))))).... ( -32.60) >consensus ___CCUCAUCGGCGACGUCAUCCGUUUCCGCUUC__________________CGCUUCCACUUCCGUGACGCCAUCGCC______AUCGCCGUUGCACCUGCAGCGGCGGCUCACAUUU ..........((((........((....))............................(((....))).))))..............(((((((((....))))))))).......... (-19.56 = -20.20 + 0.64)

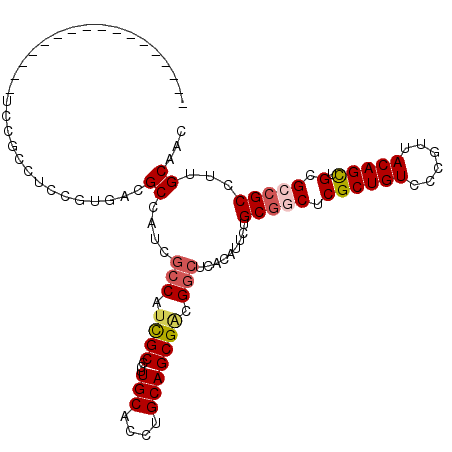
| Location | 7,091,335 – 7,091,427 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.79 |
| Mean single sequence MFE | -41.87 |
| Consensus MFE | -21.84 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7091335 92 - 23771897 AAAUGUGAGCCGCCGCUGCAGGUGCAACGGCGAU------GGCGAUGGCGUCACGGAAGUGGAAGCG------------------GAACCGGAAACGGGUGACGUCGCCGAUGAGG--- ..........(((((.(((....))).))))).(------(((((((((.((((....))))..)).------------------...(((....)))....))))))))......--- ( -42.30) >DroSec_CAF1 5979 89 - 1 AAAUGUGAGGCGCCGCUGCAGGUGCAACGGCGAU------GGCGAUGGCGUCACGGAAGUGGAAGCG------------------AAACCGGAAACGGAUGAC---GCCGAUGAGG--- ...(((.(..(((((.(((....))).))))).)------.))).(((((((((....)).......------------------...(((....)))..)))---))))......--- ( -38.00) >DroSim_CAF1 6085 89 - 1 AAAUGUGAGCCGCCGCUGCAGGUGCAACGGCGAU------GGCGAUGGCGUCACGGAAGUGGAAGCG------------------AAACCGGAAACGGAUGAC---GCCGAUGAGG--- ...(((.(..(((((.(((....))).))))).)------.))).(((((((((....)).......------------------...(((....)))..)))---))))......--- ( -37.80) >DroEre_CAF1 6468 89 - 1 AAAUGUGAGCCGCCGCUGCAGGUGCAACGGCGAG------GGCGACGGCGUCACGGAGGCG------------------------GAAGCGGAAACGGAUGUCGUCGCCGAUGUGGCGG ........(((((((.(((....))).))))...------((((((((((((......((.------------------------...))(....).)))))))))))).....))).. ( -48.00) >DroYak_CAF1 6823 113 - 1 AAAUGUGACACGGCGCUGCAGGUGCAACGGCGAU------GGCGAUGGCGUCACGGAAGCGGAUGCGGAAACGGACGCGGAACCGGAAGCGGAAACGGAUGACGUCGCCGAUGAGGCGA ...(((..((((.((.(((....))).)).)).(------(((((((.((((.(((..(((....((....))..)))....)))....((....)))))).)))))))).))..))). ( -43.50) >DroAna_CAF1 5763 95 - 1 GAAUGUCAGCCGCCGUUGCAGGUGCAAUGGCGAGGCGGCAGGCGAUGGCGUCGGCGACACG------------------------GAAGCGGAAACCGUAGACUCCUCCGUUGACGACG ...((((.(((((((((((....))))))))..((((.((.....)).)))))))))))((------------------------..((((((.............))))))..))... ( -41.62) >consensus AAAUGUGAGCCGCCGCUGCAGGUGCAACGGCGAU______GGCGAUGGCGUCACGGAAGCGGAAGCG__________________GAACCGGAAACGGAUGACGUCGCCGAUGAGG___ ..........(((((.(((....))).)))))..........((.((((..(((....)))............................((....)).........)))).))...... (-21.84 = -22.40 + 0.56)


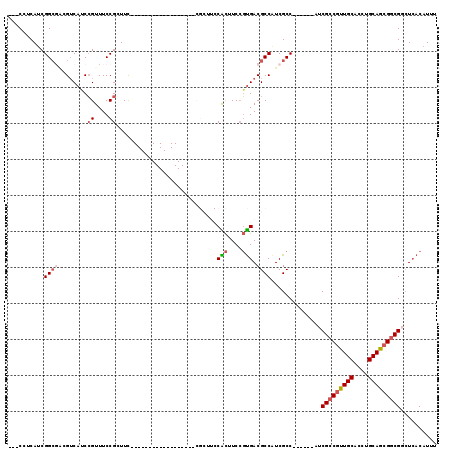
| Location | 7,091,366 – 7,091,467 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.52 |
| Mean single sequence MFE | -40.02 |
| Consensus MFE | -24.44 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7091366 101 + 23771897 -------------CGCUUCCACUUCCGUGACGCCAUCGCCAUCGCCGUUGCACCUGCAGCGGCGGCUCACAUUUUGCGGCUCGCUGUCCCGUUACAGCUGCGCCGCCUUGCAAC -------------.((..........((((.(((...(....)((((((((....))))))))))))))).....(((((.((((((......))))).).)))))...))... ( -42.20) >DroPse_CAF1 53250 97 + 1 -----------------UCCGCGUCCGUCUCGCCAGUGCCAUCGCCCCUGCAUCUGCAGCGACGGCUCACGUUCUGCGGCUCGCUGUCGCGGUACAGCUGCGGCGCCCUGCAAC -----------------...(((.((((...((..(.(((.((((...(((....))))))).))).)..((.(((((((.....))))))).)).)).)))))))........ ( -37.90) >DroEre_CAF1 6502 95 + 1 -------------------CGCCUCCGUGACGCCGUCGCCCUCGCCGUUGCACCUGCAGCGGCGGCUCACAUUUUGCGGCUCGCUGUCCCGUUACAGCUGCGCCGCCUUGCAAC -------------------.((....((((.((((.......)((((((((....))))))))))))))).....(((((.((((((......))))).).)))))...))... ( -42.50) >DroYak_CAF1 6862 114 + 1 CCGCGUCCGUUUCCGCAUCCGCUUCCGUGACGCCAUCGCCAUCGCCGUUGCACCUGCAGCGCCGUGUCACAUUUUGCGGCUCGCUGUCCCGUUACAGCUGCGCCGCCUUGCAAC ..(((........)))....((....(((((((....((....))((((((....))))))..))))))).....(((((.((((((......))))).).)))))...))... ( -40.50) >DroMoj_CAF1 32827 107 + 1 -AGAG------ACAGAGUCGGCCUCCGGGUUGCCACUGCCAUUGCCCUUGCAGCUGCAGCGACGGCUCACGUACUGCUGCUCGCUGUCCCGUUACAGUGGCGCCGCCUUGCAAC -...(------...(((.((((....((....))...(((((((.....(((((.(((((.(((.....)))...)))))..))))).......))))))))))).))).)... ( -39.10) >DroPer_CAF1 54796 97 + 1 -----------------UCCGCGUCCGUCUCGCCAGUGCCAUCGCCCCUGCAUCUGCAGCGACGGCUCACGUUCUGCGGCUCGCUGUCGCGGUACAGCUGCGGCGCCCUGCAAC -----------------...(((.((((...((..(.(((.((((...(((....))))))).))).)..((.(((((((.....))))))).)).)).)))))))........ ( -37.90) >consensus _________________UCCGCCUCCGUGACGCCAUCGCCAUCGCCCUUGCACCUGCAGCGACGGCUCACAUUCUGCGGCUCGCUGUCCCGUUACAGCUGCGCCGCCUUGCAAC ...............................((....(((.((((...(((....))))))).))).........(((((.((((((......))))).).)))))...))... (-24.44 = -25.00 + 0.56)


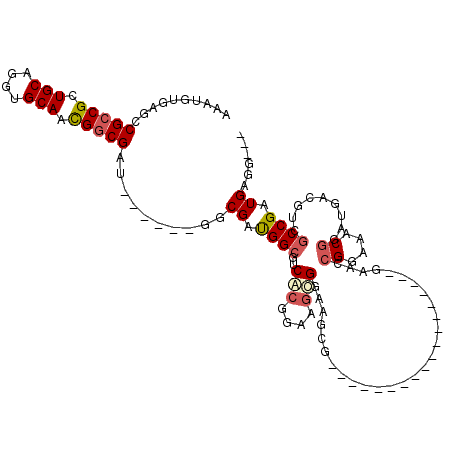
| Location | 7,091,366 – 7,091,467 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.52 |
| Mean single sequence MFE | -43.45 |
| Consensus MFE | -30.30 |
| Energy contribution | -30.72 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7091366 101 - 23771897 GUUGCAAGGCGGCGCAGCUGUAACGGGACAGCGAGCCGCAAAAUGUGAGCCGCCGCUGCAGGUGCAACGGCGAUGGCGAUGGCGUCACGGAAGUGGAAGCG------------- (((((...(((((...(((((......)))))..)))))...........(((((.(((....))).)))))...))))).((.((((....))))..)).------------- ( -45.40) >DroPse_CAF1 53250 97 - 1 GUUGCAGGGCGCCGCAGCUGUACCGCGACAGCGAGCCGCAGAACGUGAGCCGUCGCUGCAGAUGCAGGGGCGAUGGCACUGGCGAGACGGACGCGGA----------------- (((((.((...)))))))....(((((.(..(..(((((.....))..(((((((((.(.......).)))))))))...)))..)...).))))).----------------- ( -39.90) >DroEre_CAF1 6502 95 - 1 GUUGCAAGGCGGCGCAGCUGUAACGGGACAGCGAGCCGCAAAAUGUGAGCCGCCGCUGCAGGUGCAACGGCGAGGGCGACGGCGUCACGGAGGCG------------------- ...((...(((((...(((((......)))))..)))))....((((((((((((.(((....))).))))...(....)))).)))))...)).------------------- ( -44.10) >DroYak_CAF1 6862 114 - 1 GUUGCAAGGCGGCGCAGCUGUAACGGGACAGCGAGCCGCAAAAUGUGACACGGCGCUGCAGGUGCAACGGCGAUGGCGAUGGCGUCACGGAAGCGGAUGCGGAAACGGACGCGG ((..((..(((((...(((((......)))))..)))))....))..)).((.((.(((....))).)).))...(((...(((((.((....)))))))(....)...))).. ( -49.60) >DroMoj_CAF1 32827 107 - 1 GUUGCAAGGCGGCGCCACUGUAACGGGACAGCGAGCAGCAGUACGUGAGCCGUCGCUGCAGCUGCAAGGGCAAUGGCAGUGGCAACCCGGAGGCCGACUCUGU------CUCU- ((((((.(((...)))..))))))(((((((.(.(((((.(((.((((....))))))).)))))...(((..(((....(....))))...)))..).))))------))).- ( -41.80) >DroPer_CAF1 54796 97 - 1 GUUGCAGGGCGCCGCAGCUGUACCGCGACAGCGAGCCGCAGAACGUGAGCCGUCGCUGCAGAUGCAGGGGCGAUGGCACUGGCGAGACGGACGCGGA----------------- (((((.((...)))))))....(((((.(..(..(((((.....))..(((((((((.(.......).)))))))))...)))..)...).))))).----------------- ( -39.90) >consensus GUUGCAAGGCGGCGCAGCUGUAACGGGACAGCGAGCCGCAAAACGUGAGCCGUCGCUGCAGGUGCAACGGCGAUGGCAAUGGCGACACGGAAGCGGA_________________ ...((...(((((...(((((......)))))..)))))....((((...(((((.(((...(((....)))...))).))))).))))...)).................... (-30.30 = -30.72 + 0.42)


| Location | 7,091,393 – 7,091,501 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -44.53 |
| Consensus MFE | -23.59 |
| Energy contribution | -25.48 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7091393 108 - 23771897 G-AC-----UCGCUGCACACUCACUUGAGAUUCUGCACCAGUUGCAAGGCGGCGCAGCUGUAACGGGACAGCGAGCCGCAAAAUGUGAGCCGCCGCUGCAGGUGCAACGGCGAU------ .-..-----((((((....(((....)))....((((((...((((.((((((((.(((((......)))))..))(((.....))).))))))..)))))))))).)))))).------ ( -49.60) >DroVir_CAF1 35655 113 - 1 GCAUCUGUGGC-ACACAGACUCACUUGAGAUUCUGCACCAGUUGCAAGGCGGCGCCGCUGUAACGGGACAGCGCGCAACAAUACGUGACUCGGCGCUGCAGCUGCGAGGGCAGC------ ...((((((..-.))))))(((....)))...((((.((..(((((..(((((((((((((......))))).(((........)))....))))))))...))))))))))).------ ( -46.60) >DroPse_CAF1 53273 107 - 1 G-AC-----UC-CCAUCCACUCACUUGAGGUUCUGCACCAGUUGCAGGGCGCCGCAGCUGUACCGCGACAGCGAGCCGCAGAACGUGAGCCGUCGCUGCAGAUGCAGGGGCGAU------ .-.(-----((-.(.....(((....)))(((((((..(((((((.((...)))))))))...(((....)))....)))))))).))).((((.((((....)))).))))..------ ( -44.00) >DroWil_CAF1 7736 107 - 1 U-AA-----UUACUC-CAACUCACUUGAGAUUCUUCACCAAAUUCAAAGCGGAGCAACUGUAGCGGGACAAGGAGCCACAGAACGUGAGGCGUCGCUGUAGAUGCAAAGGCGAC------ .-..-----....((-(..(((....))).....((.((...(((.....)))((.......))))))...)))(((......(....)(((((......)))))...)))...------ ( -23.10) >DroAna_CAF1 5818 112 - 1 ---G-----CCGCCGCGGACUCACUUUAGGUUCUGCACCAGUUGCAAGGCGGCACAGCUGUAGCGGGACAGCGAGCCGCAGAAUGUCAGCCGCCGUUGCAGGUGCAAUGGCGAGGCGGCA ---(-----(((((((((.(((.((....(((((((..(((((((......)))..))))..))))))))).)))))))...........(((((((((....))))))))).)))))). ( -59.90) >DroPer_CAF1 54819 107 - 1 G-AC-----UC-CCAUCCACUCACUUGAGGUUCUGCACCAGUUGCAGGGCGCCGCAGCUGUACCGCGACAGCGAGCCGCAGAACGUGAGCCGUCGCUGCAGAUGCAGGGGCGAU------ .-.(-----((-.(.....(((....)))(((((((..(((((((.((...)))))))))...(((....)))....)))))))).))).((((.((((....)))).))))..------ ( -44.00) >consensus G_AC_____UC_CCACCCACUCACUUGAGAUUCUGCACCAGUUGCAAGGCGGCGCAGCUGUAACGGGACAGCGAGCCGCAGAACGUGAGCCGUCGCUGCAGAUGCAAGGGCGAU______ ...................(((((.........((((.....))))..(((((...(((((......)))))..))))).....))))).((((..(((....)))..))))........ (-23.59 = -25.48 + 1.89)



| Location | 7,091,427 – 7,091,519 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -20.48 |
| Energy contribution | -20.98 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7091427 92 - 23771897 AGAUGCAAAAGGACCUUCGACUCGCUGCACACUCACUUGAGAUUCUGCACCAGUUGCAAGGCGGCGCAGCUGUAACGGGACAGCGAGCCGCA ...(((.....(.(((((((((.(.((((..(((....)))....))))).))))).)))))(((...(((((......)))))..)))))) ( -31.40) >DroPse_CAF1 53307 78 - 1 -------------CACUCGACUC-CCAUCCACUCACUUGAGGUUCUGCACCAGUUGCAGGGCGCCGCAGCUGUACCGCGACAGCGAGCCGCA -------------..........-.......(((....)))((((((((.....))))))))((.((.(((((......)))))..)).)). ( -25.40) >DroSec_CAF1 6068 88 - 1 ----GCAAAAGGACCUUCGACCCGCUGCACACUCACUUGAGAUUCUGCACCAGUUGCAAGGCGGCGCAGCUGUAACGGGACAGCGAGCCGCA ----..........(((((((..(.((((..(((....)))....)))).).)))).)))(((((...(((((......)))))..))))). ( -29.00) >DroYak_CAF1 6936 92 - 1 GGCAGCAAAAGGACCUUCGACCUGCUGCACACUCACUUGAGAUUCUGCACCAGUUGCAAGGCGGCGCAGCUGUAACGGGACAGCGAGCCGCA .((((((...((........))))))))...(((....)))....((((.....))))..(((((...(((((......)))))..))))). ( -33.90) >DroAna_CAF1 5858 75 - 1 --------------UCU---GCCGCCGCGGACUCACUUUAGGUUCUGCACCAGUUGCAAGGCGGCACAGCUGUAGCGGGACAGCGAGCCGCA --------------...---......((((.(((.((....(((((((..(((((((......)))..))))..))))))))).))))))). ( -29.20) >DroPer_CAF1 54853 78 - 1 -------------CACUCGACUC-CCAUCCACUCACUUGAGGUUCUGCACCAGUUGCAGGGCGCCGCAGCUGUACCGCGACAGCGAGCCGCA -------------..........-.......(((....)))((((((((.....))))))))((.((.(((((......)))))..)).)). ( -25.40) >consensus _____________CCCUCGACCCGCCGCACACUCACUUGAGAUUCUGCACCAGUUGCAAGGCGGCGCAGCUGUAACGGGACAGCGAGCCGCA ...............................(((....)))....((((.....))))..(((((...(((((......)))))..))))). (-20.48 = -20.98 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:59 2006