
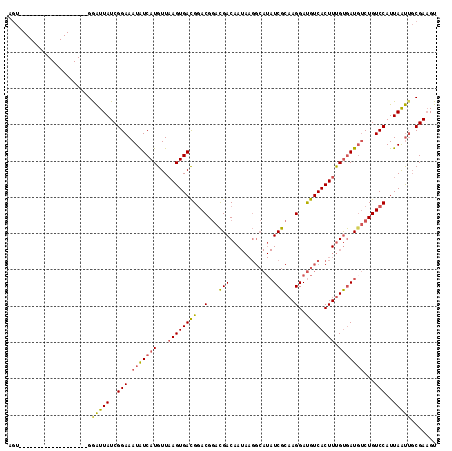
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,084,210 – 7,084,331 |
| Length | 121 |
| Max. P | 0.867221 |

| Location | 7,084,210 – 7,084,309 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.05 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -20.67 |
| Energy contribution | -20.53 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7084210 99 + 23771897 AGU-------------------GGAUUAUCGGAAAUAUCAUGUUAAGUGACGGACGGACGACAAUAAGGCAUAUCGCAAGGAUGUCACUUUGUGAUGUCUGUCCAUUAAUUGCGAAGU ...-------------------......(((.((...((((.....)))).(((((((((((((...(((((.((....)))))))...))))..))))))))).....)).)))... ( -29.30) >DroSec_CAF1 96584 99 + 1 AGU-------------------GGAUUAUCGGAAAUAUCAUGUUAAGUGACGGACGGACGACAAUAAGGCAUAUCGCAAGGAUGUCACUUUGUGAUGUCUGUCCAUUAAUUGCGAAGU ...-------------------......(((.((...((((.....)))).(((((((((((((...(((((.((....)))))))...))))..))))))))).....)).)))... ( -29.30) >DroSim_CAF1 129276 99 + 1 AGU-------------------GGAUUAUCGGAAAUAUCAUGUUAAGUGACGGACGGACGACAAUAAGGCAUAUCGCAAGGAUGUCACUUUGUGAUGUCUGUCCAUUAAUUGCGAAGU ...-------------------......(((.((...((((.....)))).(((((((((((((...(((((.((....)))))))...))))..))))))))).....)).)))... ( -29.30) >DroEre_CAF1 112765 117 + 1 AGUGGAGUCG-GCACUUUGCGGGAAUUAUCGGAAAUAUCAUGUUAAGUGAUGGACGGACGACAAUAAGGCAUAUCGCAAGGAUGUCACUUUGUGAUUUCUGUCCAUUAAUUGCGAAGU ..........-..(((((((((((....)).................(((((((((((..((((...(((((.((....)))))))...))))....))))))))))).))))))))) ( -32.20) >DroYak_CAF1 112138 118 + 1 AGUUGAGUCGGGCACUUUGCGGGAAUUAUCGGAAAUAUCAUGUUAAGUGACGGACGGACGACAAUAAGGCAUAUCGCAAGGAUGUCACUUUGUGAUGUCUGUCCAUUAAUUGCGAAGU .............(((((((((...............((((.....)))).(((((((((((((...(((((.((....)))))))...))))..))))))))).....))))))))) ( -35.00) >DroAna_CAF1 111266 88 + 1 UCU-------------------GGGUUAUCGGAAAUGUCAUGUUCAGUGACG-ACGACUGACAA---------UCGCCAGGACGUCACUUCGCAAUGCCUGUCCAUUAGC-CCGAUGU ..(-------------------((((((..(((...(.(((((..(((((((-....(((....---------....)))..)))))))..)).))).)..)))..))))-))).... ( -28.40) >consensus AGU___________________GGAUUAUCGGAAAUAUCAUGUUAAGUGACGGACGGACGACAAUAAGGCAUAUCGCAAGGAUGUCACUUUGUGAUGUCUGUCCAUUAAUUGCGAAGU .......................(((((..(((.(((((((...((((((((..(...(((............)))...)..)))))))).)))))))...)))..)))))....... (-20.67 = -20.53 + -0.13)


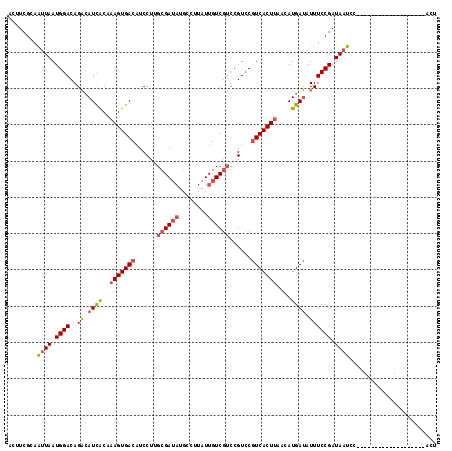
| Location | 7,084,210 – 7,084,309 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.05 |
| Mean single sequence MFE | -21.77 |
| Consensus MFE | -15.44 |
| Energy contribution | -16.08 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7084210 99 - 23771897 ACUUCGCAAUUAAUGGACAGACAUCACAAAGUGACAUCCUUGCGAUAUGCCUUAUUGUCGUCCGUCCGUCACUUAACAUGAUAUUUCCGAUAAUCC-------------------ACU ........((((.((((..((.((((..(((((((......((((((........))))))......)))))))....)))).)))))).))))..-------------------... ( -19.30) >DroSec_CAF1 96584 99 - 1 ACUUCGCAAUUAAUGGACAGACAUCACAAAGUGACAUCCUUGCGAUAUGCCUUAUUGUCGUCCGUCCGUCACUUAACAUGAUAUUUCCGAUAAUCC-------------------ACU ........((((.((((..((.((((..(((((((......((((((........))))))......)))))))....)))).)))))).))))..-------------------... ( -19.30) >DroSim_CAF1 129276 99 - 1 ACUUCGCAAUUAAUGGACAGACAUCACAAAGUGACAUCCUUGCGAUAUGCCUUAUUGUCGUCCGUCCGUCACUUAACAUGAUAUUUCCGAUAAUCC-------------------ACU ........((((.((((..((.((((..(((((((......((((((........))))))......)))))))....)))).)))))).))))..-------------------... ( -19.30) >DroEre_CAF1 112765 117 - 1 ACUUCGCAAUUAAUGGACAGAAAUCACAAAGUGACAUCCUUGCGAUAUGCCUUAUUGUCGUCCGUCCAUCACUUAACAUGAUAUUUCCGAUAAUUCCCGCAAAGUGC-CGACUCCACU ((((.(((((((.((((..((.((((..((((((.......((((((........)))))).......))))))....)))).)))))).)))))...)).))))..-.......... ( -21.24) >DroYak_CAF1 112138 118 - 1 ACUUCGCAAUUAAUGGACAGACAUCACAAAGUGACAUCCUUGCGAUAUGCCUUAUUGUCGUCCGUCCGUCACUUAACAUGAUAUUUCCGAUAAUUCCCGCAAAGUGCCCGACUCAACU ((((.(((((((.((((..((.((((..(((((((......((((((........))))))......)))))))....)))).)))))).)))))...)).))))............. ( -24.30) >DroAna_CAF1 111266 88 - 1 ACAUCGG-GCUAAUGGACAGGCAUUGCGAAGUGACGUCCUGGCGA---------UUGUCAGUCGU-CGUCACUGAACAUGACAUUUCCGAUAACCC-------------------AGA .....((-(.((.((((...((((.....(((((((.((((((..---------..)))))..).-)))))))....))).)...)))).)).)))-------------------... ( -27.20) >consensus ACUUCGCAAUUAAUGGACAGACAUCACAAAGUGACAUCCUUGCGAUAUGCCUUAUUGUCGUCCGUCCGUCACUUAACAUGAUAUUUCCGAUAAUCC___________________ACU ........((((.((((..((.((((..(((((((......((((((........))))))......)))))))....)))).)))))).))))........................ (-15.44 = -16.08 + 0.64)


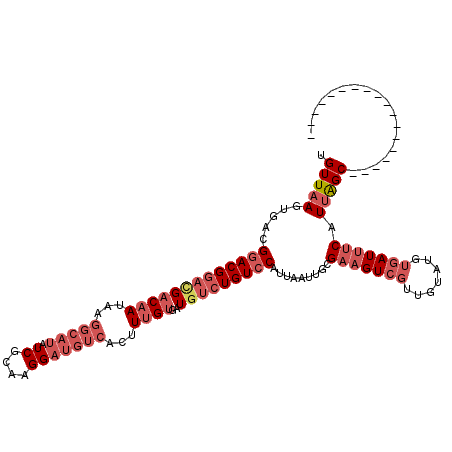
| Location | 7,084,231 – 7,084,331 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.83 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -22.26 |
| Energy contribution | -24.68 |
| Covariance contribution | 2.42 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7084231 100 + 23771897 UGUUAAGUGACGGACGGACGACAAUAAGGCAUAUCGCAAGGAUGUCACUUUGUGAUGUCUGUCCAUUAAUUGCGAAGUCGUUGUAUGUGAUUUCAUUAGC------------------ .(((((.....(((((((((((((...(((((.((....)))))))...))))..))))))))).........(((((((.......))))))).)))))------------------ ( -31.50) >DroSec_CAF1 96605 100 + 1 UGUUAAGUGACGGACGGACGACAAUAAGGCAUAUCGCAAGGAUGUCACUUUGUGAUGUCUGUCCAUUAAUUGCGAAGUCGUUGUAUGUGAUUUCAUUAGC------------------ .(((((.....(((((((((((((...(((((.((....)))))))...))))..))))))))).........(((((((.......))))))).)))))------------------ ( -31.50) >DroSim_CAF1 129297 100 + 1 UGUUAAGUGACGGACGGACGACAAUAAGGCAUAUCGCAAGGAUGUCACUUUGUGAUGUCUGUCCAUUAAUUGCGAAGUCGUUGUAUGUGAUUUCAUUAGC------------------ .(((((.....(((((((((((((...(((((.((....)))))))...))))..))))))))).........(((((((.......))))))).)))))------------------ ( -31.50) >DroEre_CAF1 112804 113 + 1 UGUUAAGUGAUGGACGGACGACAAUAAGGCAUAUCGCAAGGAUGUCACUUUGUGAUUUCUGUCCAUUAAUUGCGAAGUCGUUGUAUGUGAUUUCAUUAGCG-----CCUCCGCCGAUC .......(((((((((((..((((...(((((.((....)))))))...))))....)))))))))))...(((.((.(((((.(((......))))))))-----.)).)))..... ( -34.90) >DroYak_CAF1 112178 113 + 1 UGUUAAGUGACGGACGGACGACAAUAAGGCAUAUCGCAAGGAUGUCACUUUGUGAUGUCUGUCCAUUAAUUGCGAAGUCGUUGUAUGUGAUUUCAUUAGCG-----CCUCCGUCGAUC .......(((((((((((((((((...(((((.((....)))))))...))))..))))))))).........(.((.(((((.(((......))))))))-----.)).)))))... ( -34.40) >DroAna_CAF1 111287 107 + 1 UGUUCAGUGACG-ACGACUGACAA---------UCGCCAGGACGUCACUUCGCAAUGCCUGUCCAUUAGC-CCGAUGUGGUUUCUCCAGAUUCCAUCGGCGGCCCGCCCCAGCCUCCC (((..(((((((-....(((....---------....)))..)))))))..)))..............((-(.((((..(((......)))..)))))))(((........))).... ( -28.90) >consensus UGUUAAGUGACGGACGGACGACAAUAAGGCAUAUCGCAAGGAUGUCACUUUGUGAUGUCUGUCCAUUAAUUGCGAAGUCGUUGUAUGUGAUUUCAUUAGC__________________ .(((((.....(((((((((((((...(((((.((....)))))))...))))..))))))))).........(((((((.......))))))).))))).................. (-22.26 = -24.68 + 2.42)

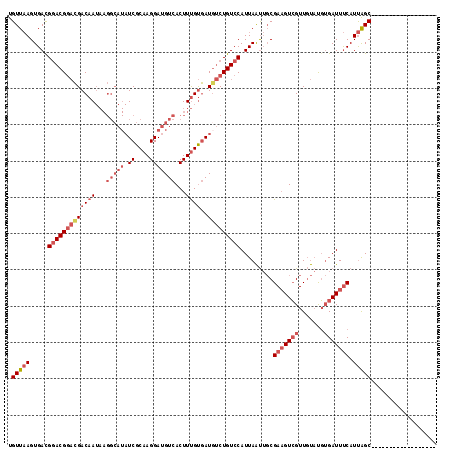
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:53 2006