| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,073,929 – 7,074,024 |
| Length | 95 |
| Max. P | 0.669587 |

| Location | 7,073,929 – 7,074,024 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.20 |
| Mean single sequence MFE | -34.63 |
| Consensus MFE | -22.46 |
| Energy contribution | -22.77 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
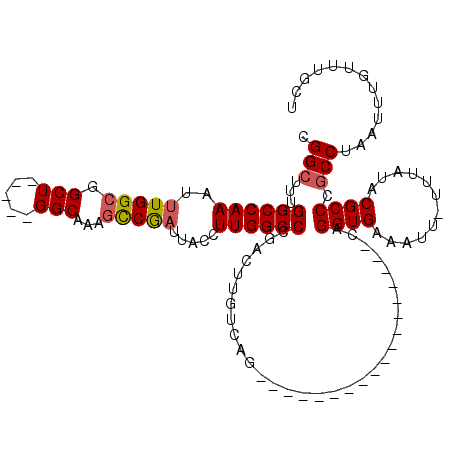
>3L_DroMel_CAF1 7073929 95 + 23771897 CGGCUUUGGCCAAAUUUGGCGGCU-----GGCGAAGCCGAUUACCUUGGCCCGACUUGUCAG------------------CAGGUGAAAUU-UUUAUACGCCCGCCUAAUUUGUUUGCU .(((....)))(((((.(((((((-----(((((.(((((.....))))).....)))))))------------------).((((.....-......)))))))).)))))....... ( -32.30) >DroSec_CAF1 86455 95 + 1 CGGCUUUGGCCAAAUUUGGCGGCU-----GGCAAAGCCGAUUACCUUGGCCCGACUUGUCAG------------------CAGGUGAAAUU-UUUAUACGCCCGCCUAAUUUGUUUGCU .(((....)))(((((.(((((((-----(((((.(((((.....))))).....)))))))------------------).((((.....-......)))))))).)))))....... ( -32.60) >DroSim_CAF1 119147 95 + 1 CGGCUUUGGCCAAAUUUGGCGGCU-----GGCAAAGCCGAUUACCUUGGCCCGACUUGUCAG------------------CAGGUGAAAUU-UUUAUACGCCCGCCUAAUUUGUUUGCU .(((....)))(((((.(((((((-----(((((.(((((.....))))).....)))))))------------------).((((.....-......)))))))).)))))....... ( -32.60) >DroEre_CAF1 102664 95 + 1 CGGCUUUGGCCAAAUUUGGCGGCU-----GGCGAAGCCGAUUACCUUGGCCAGAGUUGUCAG------------------CAGGUGAAAUU-UGUAUACGCCCGCCUAAUUUGUUUGCU (((((((((((((......(((((-----.....)))))......)))))))))))))..((------------------((((..((.(.-.((........))..).))..)))))) ( -35.00) >DroYak_CAF1 101473 100 + 1 CGGCUUUGGCCAAAUUUGGCGGCUGAUUUGGCAAAGCCGAUUACCUUGGCCCGACUUGUCAG------------------CAGGUGAAAUU-UGUAUGCGCCCGCCUAAUUUGUUUGCU .(((....)))(((((.((((((((((((((....(((((.....)))))))))...)))))------------------).((((.....-......)))))))).)))))....... ( -32.50) >DroAna_CAF1 102450 109 + 1 UGG-UCGGGCCAAACUUGGGGG---------CAGAGUCACUUGCCUUGGCCUGCCGUGCCAGCUUGCCAGCUUGCCAGCGCAGGUGAAAUUUUUUAUACGCCCGCCUUGUUUGUCUCUC ...-..(((.(((((..(((((---------(....((((((((.(((((..((.(.((......))).))..))))).))))))))((....))....)))).))..))))).))).. ( -42.80) >consensus CGGCUUUGGCCAAAUUUGGCGGCU_____GGCAAAGCCGAUUACCUUGGCCCGACUUGUCAG__________________CAGGUGAAAUU_UUUAUACGCCCGCCUAAUUUGUUUGCU .(((...((((((..(((((.(((.....)))...))))).....))))))...............................((((............)))).)))............. (-22.46 = -22.77 + 0.31)

| Location | 7,073,929 – 7,074,024 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.20 |
| Mean single sequence MFE | -29.99 |
| Consensus MFE | -20.63 |
| Energy contribution | -21.47 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
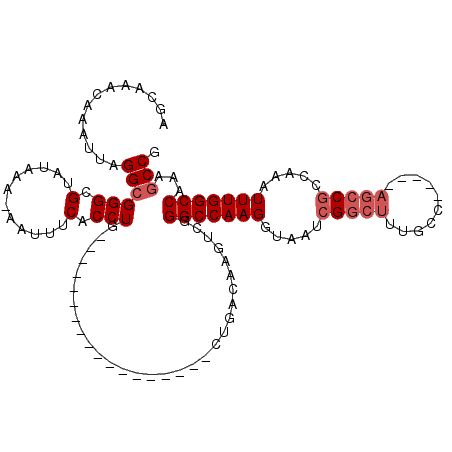
>3L_DroMel_CAF1 7073929 95 - 23771897 AGCAAACAAAUUAGGCGGGCGUAUAAA-AAUUUCACCUG------------------CUGACAAGUCGGGCCAAGGUAAUCGGCUUCGCC-----AGCCGCCAAAUUUGGCCAAAGCCG .((..........((((((.(......-.....).))))------------------)).........(((((((.....(((((.....-----))))).....)))))))...)).. ( -28.70) >DroSec_CAF1 86455 95 - 1 AGCAAACAAAUUAGGCGGGCGUAUAAA-AAUUUCACCUG------------------CUGACAAGUCGGGCCAAGGUAAUCGGCUUUGCC-----AGCCGCCAAAUUUGGCCAAAGCCG .((..........((((((.(......-.....).))))------------------)).........(((((((.....(((((.....-----))))).....)))))))...)).. ( -28.70) >DroSim_CAF1 119147 95 - 1 AGCAAACAAAUUAGGCGGGCGUAUAAA-AAUUUCACCUG------------------CUGACAAGUCGGGCCAAGGUAAUCGGCUUUGCC-----AGCCGCCAAAUUUGGCCAAAGCCG .((..........((((((.(......-.....).))))------------------)).........(((((((.....(((((.....-----))))).....)))))))...)).. ( -28.70) >DroEre_CAF1 102664 95 - 1 AGCAAACAAAUUAGGCGGGCGUAUACA-AAUUUCACCUG------------------CUGACAACUCUGGCCAAGGUAAUCGGCUUCGCC-----AGCCGCCAAAUUUGGCCAAAGCCG .((..........((((((.(......-.....).))))------------------))........((((((((.....(((((.....-----))))).....))))))))..)).. ( -29.50) >DroYak_CAF1 101473 100 - 1 AGCAAACAAAUUAGGCGGGCGCAUACA-AAUUUCACCUG------------------CUGACAAGUCGGGCCAAGGUAAUCGGCUUUGCCAAAUCAGCCGCCAAAUUUGGCCAAAGCCG .((..........((((((.(.(....-....)).))))------------------)).........(((((((.....(((((..........))))).....)))))))...)).. ( -27.00) >DroAna_CAF1 102450 109 - 1 GAGAGACAAACAAGGCGGGCGUAUAAAAAAUUUCACCUGCGCUGGCAAGCUGGCAAGCUGGCACGGCAGGCCAAGGCAAGUGACUCUG---------CCCCCAAGUUUGGCCCGA-CCA ......(((((..((.(((((...........((((.(((..((((..((((((......)).))))..))))..))).))))...))---------)))))..)))))......-... ( -37.34) >consensus AGCAAACAAAUUAGGCGGGCGUAUAAA_AAUUUCACCUG__________________CUGACAAGUCGGGCCAAGGUAAUCGGCUUUGCC_____AGCCGCCAAAUUUGGCCAAAGCCG .............((((((.(............).)))..............................(((((((.....(((((..........))))).....)))))))...))). (-20.63 = -21.47 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:45 2006