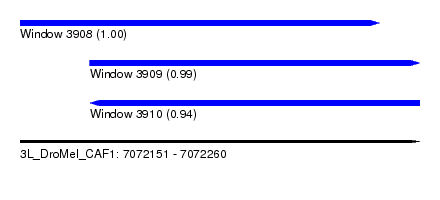
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,072,151 – 7,072,260 |
| Length | 109 |
| Max. P | 0.995525 |

| Location | 7,072,151 – 7,072,249 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 93.69 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -26.48 |
| Energy contribution | -27.28 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7072151 98 + 23771897 AAUGGCUGCUUAAAUGCGAGAAUAAUUGUUAUGCAGCAGCUG-ACGGGAGGAUGCGCAGUUGUCACUUGGCAACUGCUAUAUGAUUCUUUGCGCUUUUU ...(((((((.....((((......)))).....))))))).-....(((((.((((((..((((..((((....))))..))))...))))))))))) ( -32.00) >DroSec_CAF1 84687 98 + 1 AAUGGCUGCUUAAAUGCGCGAAUAAUUGUUAUGCAGCAGCUG-ACGGGAGGAUGCGCAGUUGUCACUUGGCAACUGCCAUAUGAUUCUUUGCGCUUUUU ...(((((((...(((.((((....)))))))..))))))).-....(((((.((((((..((((..((((....))))..))))...))))))))))) ( -32.70) >DroSim_CAF1 117383 98 + 1 AAUGGCUGCUUAAAUGCGCGAAUAAUUGUUAUGCAGCAGCUG-ACGGGAGGAUGCGCAGUUGUCACUUGGCAACUGCUAUAUGAUUCUUUGCGCUUUUU ...(((((((...(((.((((....)))))))..))))))).-....(((((.((((((..((((..((((....))))..))))...))))))))))) ( -30.40) >DroEre_CAF1 100667 98 + 1 AAUGGCUGCG-AAAUGCGAGAAUAAUUGUUAUGCAUCAGCCGAACGGGAGGAUGCGCAGUUGUCACUUGGCAACUGCUAUUUUAUUCUUUACACUUUUU ....((....-....))((((((((.......(((((..((.....))..)))))(((((((((....)))))))))....)))))))).......... ( -32.30) >DroYak_CAF1 99393 97 + 1 AAUGGCAGCG-AAAUGCGAGAAUAAUUGUUAUGCAGCAGCUG-ACGGGAGGAUGCGCAGUUGUCACUUGGCAACUGCCAUUUUAUUCUUUGCACUUUUU ....(((((.-....))((((((((((((((.((....))))-))))...((((.(((((((((....))))))))))))))))))))))))....... ( -31.10) >consensus AAUGGCUGCUUAAAUGCGAGAAUAAUUGUUAUGCAGCAGCUG_ACGGGAGGAUGCGCAGUUGUCACUUGGCAACUGCUAUAUGAUUCUUUGCGCUUUUU ...(((((((.....((((......)))).....)))))))....(((((.(((.(((((((((....))))))))))))....))))).......... (-26.48 = -27.28 + 0.80)

| Location | 7,072,170 – 7,072,260 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 94.69 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -24.06 |
| Energy contribution | -24.46 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7072170 90 + 23771897 GAAUAAUUGUUAUGCAGCAGCUG-ACGGGAGGAUGCGCAGUUGUCACUUGGCAACUGCUAUAUGAUUCUUUGCGCUUUUUGCCGCGUUUCA (((.....((((.((....))))-))((.((((.((((((..((((..((((....))))..))))...)))))).)))).))....))). ( -28.90) >DroSec_CAF1 84706 90 + 1 GAAUAAUUGUUAUGCAGCAGCUG-ACGGGAGGAUGCGCAGUUGUCACUUGGCAACUGCCAUAUGAUUCUUUGCGCUUUUUGCCGCAUUUCA (((.....((((.((....))))-))((.((((.((((((..((((..((((....))))..))))...)))))).)))).))....))). ( -31.20) >DroSim_CAF1 117402 90 + 1 GAAUAAUUGUUAUGCAGCAGCUG-ACGGGAGGAUGCGCAGUUGUCACUUGGCAACUGCUAUAUGAUUCUUUGCGCUUUUUGCCGCAUUUCA (((.....((((.((....))))-))((.((((.((((((..((((..((((....))))..))))...)))))).)))).))....))). ( -28.90) >DroEre_CAF1 100685 91 + 1 GAAUAAUUGUUAUGCAUCAGCCGAACGGGAGGAUGCGCAGUUGUCACUUGGCAACUGCUAUUUUAUUCUUUACACUUUUUGCCGCUUUUCA ((((((.......(((((..((.....))..)))))(((((((((....)))))))))....))))))....................... ( -28.70) >DroYak_CAF1 99411 90 + 1 GAAUAAUUGUUAUGCAGCAGCUG-ACGGGAGGAUGCGCAGUUGUCACUUGGCAACUGCCAUUUUAUUCUUUGCACUUUUUGCCGCGUUUCA .......((..((((.((((.((-..((((.((((.(((((((((....)))))))))))))...))))...))....)))).))))..)) ( -27.80) >consensus GAAUAAUUGUUAUGCAGCAGCUG_ACGGGAGGAUGCGCAGUUGUCACUUGGCAACUGCUAUAUGAUUCUUUGCGCUUUUUGCCGCAUUUCA .......((..((((.((((.....((.(((((((.(((((((((....)))))))))))......))))).))....)))).))))..)) (-24.06 = -24.46 + 0.40)



| Location | 7,072,170 – 7,072,260 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 94.69 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -18.78 |
| Energy contribution | -18.98 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7072170 90 - 23771897 UGAAACGCGGCAAAAAGCGCAAAGAAUCAUAUAGCAGUUGCCAAGUGACAACUGCGCAUCCUCCCGU-CAGCUGCUGCAUAACAAUUAUUC ......((((((.....................(((((((.(....).)))))))((..........-..))))))))............. ( -20.40) >DroSec_CAF1 84706 90 - 1 UGAAAUGCGGCAAAAAGCGCAAAGAAUCAUAUGGCAGUUGCCAAGUGACAACUGCGCAUCCUCCCGU-CAGCUGCUGCAUAACAAUUAUUC ....((((((((....(((((.....((((.((((....)))).))))....)))))........(.-...)))))))))........... ( -28.10) >DroSim_CAF1 117402 90 - 1 UGAAAUGCGGCAAAAAGCGCAAAGAAUCAUAUAGCAGUUGCCAAGUGACAACUGCGCAUCCUCCCGU-CAGCUGCUGCAUAACAAUUAUUC ....((((((((.....................(((((((.(....).)))))))((..........-..))))))))))........... ( -22.10) >DroEre_CAF1 100685 91 - 1 UGAAAAGCGGCAAAAAGUGUAAAGAAUAAAAUAGCAGUUGCCAAGUGACAACUGCGCAUCCUCCCGUUCGGCUGAUGCAUAACAAUUAUUC .......................((((((....(((((((.(....).)))))))(((((..((.....))..))))).......)))))) ( -21.70) >DroYak_CAF1 99411 90 - 1 UGAAACGCGGCAAAAAGUGCAAAGAAUAAAAUGGCAGUUGCCAAGUGACAACUGCGCAUCCUCCCGU-CAGCUGCUGCAUAACAAUUAUUC ......((((((....((((...........((((....))))(((....)))))))........(.-...)))))))............. ( -20.80) >consensus UGAAACGCGGCAAAAAGCGCAAAGAAUCAUAUAGCAGUUGCCAAGUGACAACUGCGCAUCCUCCCGU_CAGCUGCUGCAUAACAAUUAUUC ......((((((.....................(((((((.(....).)))))))((.............))))))))............. (-18.78 = -18.98 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:40 2006