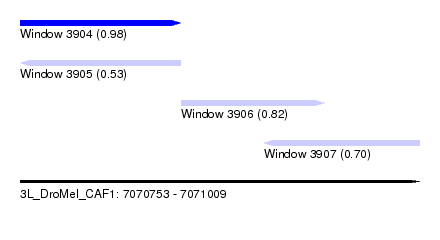
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,070,753 – 7,071,009 |
| Length | 256 |
| Max. P | 0.979940 |

| Location | 7,070,753 – 7,070,856 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 90.19 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -25.58 |
| Energy contribution | -25.14 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
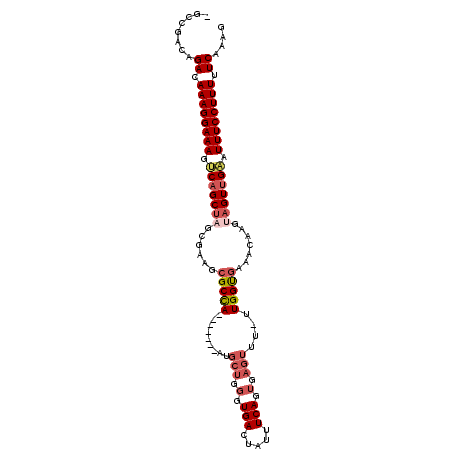
>3L_DroMel_CAF1 7070753 103 + 23771897 UAUAGUAGAUAUA----UAUAUAUCGCGAAUAGCACUGCUGGCUGGCUGCAAAGUCAAUUGCCAAAUAUGGCGGACUUGUUUGUGUGUGGUUUAAUGUCACACUCAC---- .......(((((.----...)))))(((..((((.(....)))))..))).(((((....((((....)))).)))))...((.((((((.......)))))).)).---- ( -27.10) >DroSec_CAF1 83329 101 + 1 UAUAGUAGAUAU------AUAUAUUGCGAAUAGCACUGCUGGCUGGCUGCAAAGUCAAUUGCCAAAUAUGGCGGACUUGUUUGUGUGUGGUUUAAUGUCACACUCAC---- ............------......((((..((((.(....)))))..))))(((((....((((....)))).)))))...((.((((((.......)))))).)).---- ( -25.00) >DroSim_CAF1 116015 107 + 1 UAUAGUAGAUAUAUGUAUAUAUAUUGCGAAUAGCACUGCUGGCUGGCUGCAAAGUCAAUUGCCAAAUAUGGCGGACUUGUUUGUGUGUGGUUUAAUGUCACACUCAC---- ..((((((((((((....))))))(((.....)))))))))((.....)).(((((....((((....)))).)))))...((.((((((.......)))))).)).---- ( -27.20) >DroEre_CAF1 99348 95 + 1 UAUAGCAGAUAUA--------UAUCGCGAAUAGCACUGCUAGCUGGCUGCAAAGUCAAUUGCCAAAUAUGGCAGACUUGUUUGUGUGUGGCUUAAUGUCACAC-------- ....((((((...--------....(((..((((.......))))..))).(((((...(((((....))))))))))))))))(((((((.....)))))))-------- ( -30.70) >DroYak_CAF1 97932 106 + 1 UAUAGCAGAUAUA----CA-AUAUCGCGAAUAGCACUGCUGCCUGGCUGCAAAGUCAAUUGCCAAAUAUGGCAGACUUGUUUGUGUGUGGUUUAAUGUCACACUCACACAU ..((((((((((.----..-)))))((.....))..)))))..(((((....))))).((((((....))))))...(((.((.((((((.......)))))).)).))). ( -29.40) >consensus UAUAGUAGAUAUA_____AUAUAUCGCGAAUAGCACUGCUGGCUGGCUGCAAAGUCAAUUGCCAAAUAUGGCGGACUUGUUUGUGUGUGGUUUAAUGUCACACUCAC____ .......(((((........)))))((((((.(((..(((....)))))).(((((...(((((....))))))))))))))))(((((((.....)))))))........ (-25.58 = -25.14 + -0.44)


| Location | 7,070,753 – 7,070,856 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 90.19 |
| Mean single sequence MFE | -22.98 |
| Consensus MFE | -17.54 |
| Energy contribution | -18.34 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7070753 103 - 23771897 ----GUGAGUGUGACAUUAAACCACACACAAACAAGUCCGCCAUAUUUGGCAAUUGACUUUGCAGCCAGCCAGCAGUGCUAUUCGCGAUAUAUA----UAUAUCUACUAUA ----(((((((((.........)))))......(((((.((((....))))....))))).(((((......))..)))...))))(((((...----.)))))....... ( -22.30) >DroSec_CAF1 83329 101 - 1 ----GUGAGUGUGACAUUAAACCACACACAAACAAGUCCGCCAUAUUUGGCAAUUGACUUUGCAGCCAGCCAGCAGUGCUAUUCGCAAUAUAU------AUAUCUACUAUA ----(((((((((.........)))))......(((((.((((....))))....))))).(((((......))..)))...)))).......------............ ( -20.00) >DroSim_CAF1 116015 107 - 1 ----GUGAGUGUGACAUUAAACCACACACAAACAAGUCCGCCAUAUUUGGCAAUUGACUUUGCAGCCAGCCAGCAGUGCUAUUCGCAAUAUAUAUACAUAUAUCUACUAUA ----(((((((((.........)))))......(((((.((((....))))....))))).(((((......))..)))...)))).((((((....))))))........ ( -20.50) >DroEre_CAF1 99348 95 - 1 --------GUGUGACAUUAAGCCACACACAAACAAGUCUGCCAUAUUUGGCAAUUGACUUUGCAGCCAGCUAGCAGUGCUAUUCGCGAUA--------UAUAUCUGCUAUA --------(((((.(.....).)))))......((((((((((....)))))...))))).((.....))(((((((((.....)))...--------.....)))))).. ( -23.10) >DroYak_CAF1 97932 106 - 1 AUGUGUGAGUGUGACAUUAAACCACACACAAACAAGUCUGCCAUAUUUGGCAAUUGACUUUGCAGCCAGGCAGCAGUGCUAUUCGCGAUAU-UG----UAUAUCUGCUAUA .((((((.(.((........))).))))))...((((((((((....)))))...))))).((((....((((...(((.....)))...)-))----)....)))).... ( -29.00) >consensus ____GUGAGUGUGACAUUAAACCACACACAAACAAGUCCGCCAUAUUUGGCAAUUGACUUUGCAGCCAGCCAGCAGUGCUAUUCGCGAUAUAU_____UAUAUCUACUAUA ....(((((((((.........)))))......(((((.((((....))))....))))).(((((......))..)))...))))......................... (-17.54 = -18.34 + 0.80)

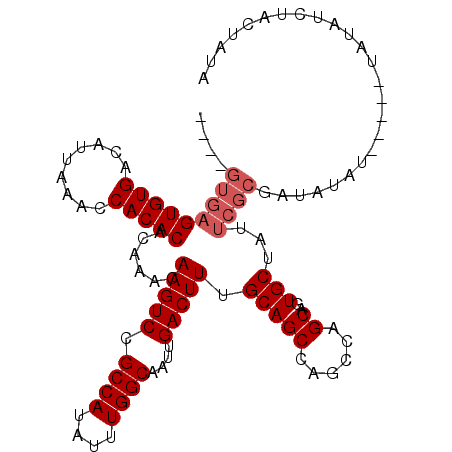

| Location | 7,070,856 – 7,070,948 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.72 |
| Mean single sequence MFE | -18.37 |
| Consensus MFE | -16.07 |
| Energy contribution | -15.58 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.822060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7070856 92 + 23771897 ----------A-----------CACUAACACCAGCCCA-----GAGAUAAGUAUAACUCUCUGCCUGUCAAUAGCUGGGCUUUGAAAAAGGAAAUUCAACUACUUGUUUCACCAA-AAA ----------.-----------................-----((((((((((..........(((.((((.(((...)))))))...))).........)))))))))).....-... ( -17.61) >DroSec_CAF1 83430 92 + 1 ----------A-----------CACUAACACCAGCCCA-----GAGAUAAGUAUAACUCUCUGUCUGUCAAUAGCUGGGCUUUGAAAAAGGAAAUUCAACUACUUGUUUCACCAA-AAA ----------.-----------..........((((((-----(.((((..((........))..)))).....))))))).((((((((............))).)))))....-... ( -15.90) >DroSim_CAF1 116122 92 + 1 ----------A-----------CACUAACACCAGCCCA-----GAGAUAAGUAUAACUCUCUGCCUGUCAAUAGCUGGGCUUUGAAAAAGGAAAUUCAACUACUUGUUUCACCAA-AAA ----------.-----------................-----((((((((((..........(((.((((.(((...)))))))...))).........)))))))))).....-... ( -17.61) >DroEre_CAF1 99443 98 + 1 ----ACACUCA-----------CACUCACACCAGCCCA-----GAGAUAAGUAUAACUCUCUGCCUGUCAAUAGCUGAGCUUUGAAAAAGGAAAUUCAACUACUUGUUUCACCAA-AAA ----.......-----------................-----((((((((((..........(((.((((.(((...)))))))...))).........)))))))))).....-... ( -17.61) >DroYak_CAF1 98038 113 + 1 CUACACACUCACACACUAAG-ACACUCACACCAUCGCA-----GAGAUAAGUAUAACUCUCUGGCUGUCAAUAGCUGAGCUUUGAAAAAGGAAAUUCAACUACUUGUUUCACCAAAAAA ....................-.................-----((((((((((.....(((.(((((....))))))))((((....)))).........))))))))))......... ( -18.80) >DroAna_CAF1 99924 110 + 1 CAGCACU-------UCUAUUGACACUAGCACUAGCUCACUGUGAAGAUAAGUGUAACUCUCUGCCUGUCAAUGGCUGAGCUUUGAAAAAGGAAAUACCACUACUUGUUUUGCCAA-A-A ..((...-------........(((.(((....)))....)))((((((((((.....(((.(((.......))).)))((((....)))).........))))))))))))...-.-. ( -22.70) >consensus __________A___________CACUAACACCAGCCCA_____GAGAUAAGUAUAACUCUCUGCCUGUCAAUAGCUGAGCUUUGAAAAAGGAAAUUCAACUACUUGUUUCACCAA_AAA ...........................................((((((((((..........(((.((((.(((...)))))))...))).........))))))))))......... (-16.07 = -15.58 + -0.50)

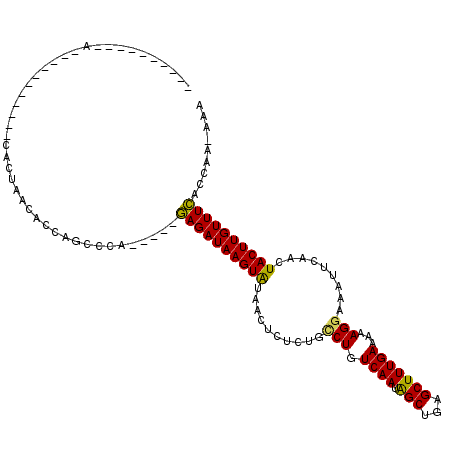

| Location | 7,070,909 – 7,071,009 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 85.89 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -23.20 |
| Energy contribution | -24.37 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7070909 100 - 23771897 -GCCGACAGACAAAGGAAAGUCAGCUAGCGAAGCGCCA-------AUGCUGGGUGACUAUUUCAGUGAGUUU-UUGGUGAAACAAGUAGUUGAAUUUCCUUUUUCAAAG -.......((.((((((((.(((((((......(((((-------(.(((.(.(((.....))).).)))..-)))))).......))))))).)))))))).)).... ( -32.42) >DroSec_CAF1 83483 100 - 1 -GCCGACAGACAAAGGAAAGUCAGCUAGCGAAGCGCCA-------AUGCUGGGUGACUAUUUCAGUGAGUUU-UUGGUGAAACAAGUAGUUGAAUUUCCUUUUUCAAAG -.......((.((((((((.(((((((......(((((-------(.(((.(.(((.....))).).)))..-)))))).......))))))).)))))))).)).... ( -32.42) >DroSim_CAF1 116175 100 - 1 -GCCGAUAGACAAAGGAAAGUCAGCUAGCGAAGCGCCA-------AUGCUGGGUGACUAUUUCAGUGAGUUU-UUGGUGAAACAAGUAGUUGAAUUUCCUUUUUCAAAG -.......((.((((((((.(((((((......(((((-------(.(((.(.(((.....))).).)))..-)))))).......))))))).)))))))).)).... ( -32.42) >DroEre_CAF1 99502 100 - 1 -GCCGAUAGACAAAGGAAAGUCAGCUUGCGAAGCGCCA-------AUGCUGGGUGACUAUUUCACUGAGUUU-UUGGUGAAACAAGUAGUUGAAUUUCCUUUUUCAAAG -.......((.((((((((.((((((.((....(((((-------(.(((.(((((.....))))).)))..-))))))......)))))))).)))))))).)).... ( -36.30) >DroYak_CAF1 98111 109 - 1 AGACGAGAGACAAAGGAAAGCCAGCUAGCGAAGCGCUAGUACUGAGUGCUGGGUGACUAUUUCAGUGAGUUUUUUGGUGAAACAAGUAGUUGAAUUUCCUUUUUCAAAG ........((.((((((((..(((((((((...)))...(((((((...(((....))).))))))).(((((.....)))))...))))))..)))))))).)).... ( -31.80) >DroAna_CAF1 99996 86 - 1 -GUUGCGAGACAAAGGAAAGGCAGCCCA------GCCA--------------GUGACUAUUUCAGCGACU-U-UUGGCAAAACAAGUAGUGGUAUUUCCUUUUUCAAAG -.......((.(((((((((((......------))).--------------............((.(((-(-(((......)))).))).)).)))))))).)).... ( -21.10) >consensus _GCCGACAGACAAAGGAAAGUCAGCUAGCGAAGCGCCA_______AUGCUGGGUGACUAUUUCAGUGAGUUU_UUGGUGAAACAAGUAGUUGAAUUUCCUUUUUCAAAG ........((.((((((((.(((((((......(((((.........(((.(.(((.....))).).)))....))))).......))))))).)))))))).)).... (-23.20 = -24.37 + 1.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:38 2006