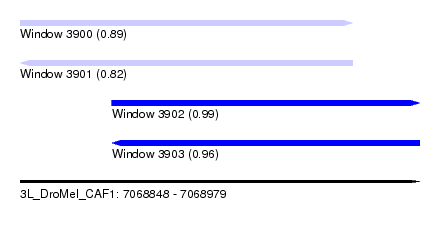
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,068,848 – 7,068,979 |
| Length | 131 |
| Max. P | 0.990791 |

| Location | 7,068,848 – 7,068,957 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.19 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -31.18 |
| Energy contribution | -32.34 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
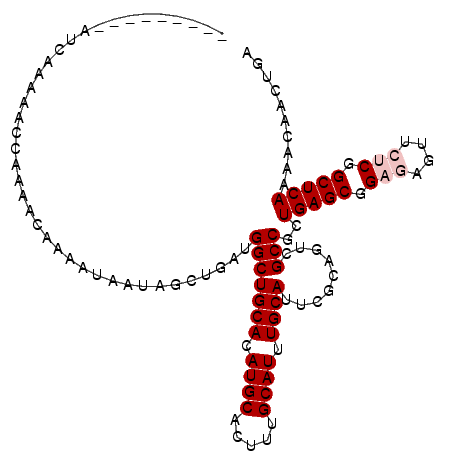
>3L_DroMel_CAF1 7068848 109 + 23771897 CGUGGUGUUUAGUGU----------UGUUUCGCGUUGUUUUCAGUUGUUUUGAGCCGAGAACUCUCCGCUCAGCGGCGACUGCGAAUGCAAAUGCAAAGUGCAUGUGCAGCCAACAGCU- .............((----------((((..((...((((.((((((((((((((.(((....))).)))))).)))))))).))))(((.((((.....)))).))).)).)))))).- ( -38.70) >DroSec_CAF1 81427 110 + 1 CGUGGUGUUUAGUGU----------UGUUUCGCGCUGUUUUCAGUUGUUUUGAGCCGAGAACUCUCCGCUCAGCGGCGACUGCGAAUGCAAAUGCAAAGUGCAUGUGCAGCCAUCAGCUA .(((((...((((((----------......))))))(((.((((((((((((((.(((....))).)))))).)))))))).)))((((.((((.....)))).)))))))))...... ( -39.90) >DroSim_CAF1 114159 110 + 1 CGUGGUGUUUAGUGU----------UGUUUCGCGCUGUUUUCAGUUGUUUUGAGCCGAGAACUCUCCGCUCAGCGGCGACUGCGAAUGCAAAUGCAAAGUGCAUGUGCAGCCAUCAGCUA .(((((...((((((----------......))))))(((.((((((((((((((.(((....))).)))))).)))))))).)))((((.((((.....)))).)))))))))...... ( -39.90) >DroEre_CAF1 97699 108 + 1 CGUGGUGUUUAGUGU----------UGUUUCGCGUUGUUUUCAGUUGCUUUGAGCCGCGAACU--CCGCUCAGCGGCAAUUGCAAAUGCAAAUGCAAAGUGCAUGUGCAGCCACAAGCUA .((((((((.((((.----------.(((.((((..((..((((.....))))))))))))).--.)))).))).(((..((((..(((....)))...))))..))).)))))...... ( -35.10) >DroYak_CAF1 95970 113 + 1 CGUGGUGUUUAGUGUUGUUCGGUGUUGUUUCGCGUUGUUUUCAGUUGCUUUGAGCCGAGAACU--CCGCUCAGCGGCAA-----AUUGCAAAUGCAAAGUGCAUGUGCAGCCAUCACCUA .((((((...............(((((((..(((..((((((.(((......))).)))))).--.)))..))))))).-----.(((((.((((.....)))).))))).))))))... ( -35.40) >consensus CGUGGUGUUUAGUGU__________UGUUUCGCGUUGUUUUCAGUUGUUUUGAGCCGAGAACUCUCCGCUCAGCGGCGACUGCGAAUGCAAAUGCAAAGUGCAUGUGCAGCCAUCAGCUA .(((((..............................((((.((((((((((((((.(((....))).)))))).)))))))).))))(((.((((.....)))).))).)))))...... (-31.18 = -32.34 + 1.16)



| Location | 7,068,848 – 7,068,957 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.19 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -24.44 |
| Energy contribution | -25.12 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7068848 109 - 23771897 -AGCUGUUGGCUGCACAUGCACUUUGCAUUUGCAUUCGCAGUCGCCGCUGAGCGGAGAGUUCUCGGCUCAAAACAACUGAAAACAACGCGAAACA----------ACACUAAACACCACG -((.(((((..((((.((((.....)))).))))((((((((....)))((((.(((....))).))))..................))))).))----------))))).......... ( -32.80) >DroSec_CAF1 81427 110 - 1 UAGCUGAUGGCUGCACAUGCACUUUGCAUUUGCAUUCGCAGUCGCCGCUGAGCGGAGAGUUCUCGGCUCAAAACAACUGAAAACAGCGCGAAACA----------ACACUAAACACCACG ..((((.((..((((.((((.....)))).))))..))))))...((((((((.(((....))).)))))......(((....))).))).....----------............... ( -32.80) >DroSim_CAF1 114159 110 - 1 UAGCUGAUGGCUGCACAUGCACUUUGCAUUUGCAUUCGCAGUCGCCGCUGAGCGGAGAGUUCUCGGCUCAAAACAACUGAAAACAGCGCGAAACA----------ACACUAAACACCACG ..((((.((..((((.((((.....)))).))))..))))))...((((((((.(((....))).)))))......(((....))).))).....----------............... ( -32.80) >DroEre_CAF1 97699 108 - 1 UAGCUUGUGGCUGCACAUGCACUUUGCAUUUGCAUUUGCAAUUGCCGCUGAGCGG--AGUUCGCGGCUCAAAGCAACUGAAAACAACGCGAAACA----------ACACUAAACACCACG ......((((.((((.((((.....)))).))))(((((....(((((.((((..--.)))))))))(((.......))).......)))))...----------..........)))). ( -31.10) >DroYak_CAF1 95970 113 - 1 UAGGUGAUGGCUGCACAUGCACUUUGCAUUUGCAAU-----UUGCCGCUGAGCGG--AGUUCUCGGCUCAAAGCAACUGAAAACAACGCGAAACAACACCGAACAACACUAAACACCACG ..((((..(((((((.((((.....)))).))))..-----..)))(((((((.(--(....)).))))..)))......................)))).................... ( -31.00) >consensus UAGCUGAUGGCUGCACAUGCACUUUGCAUUUGCAUUCGCAGUCGCCGCUGAGCGGAGAGUUCUCGGCUCAAAACAACUGAAAACAACGCGAAACA__________ACACUAAACACCACG .......(((.((((.((((.....)))).))))(((((....((((..((((.....)))).))))(((.......))).......))))).......................))).. (-24.44 = -25.12 + 0.68)


| Location | 7,068,878 – 7,068,979 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 87.43 |
| Mean single sequence MFE | -31.64 |
| Consensus MFE | -27.40 |
| Energy contribution | -28.40 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7068878 101 + 23771897 UCAGUUGUUUUGAGCCGAGAACUCUCCGCUCAGCGGCGACUGCGAAUGCAAAUGCAAAGUGCAUGUGCAGCCAACAGCU-UUAUUUUGUUUUGGUUUUUGAU--------- .((((((((((((((.(((....))).)))))).))))))))((((((((.((((.....)))).))))(((((.(((.-.......)))))))).))))..--------- ( -35.70) >DroSec_CAF1 81457 102 + 1 UCAGUUGUUUUGAGCCGAGAACUCUCCGCUCAGCGGCGACUGCGAAUGCAAAUGCAAAGUGCAUGUGCAGCCAUCAGCUAUUAUUUUGUUUUGGUUUUUGAU--------- .((((((((((((((.(((....))).)))))).))))))))((((((((.((((.....)))).))))((((..(((.........))).)))).))))..--------- ( -33.70) >DroSim_CAF1 114189 102 + 1 UCAGUUGUUUUGAGCCGAGAACUCUCCGCUCAGCGGCGACUGCGAAUGCAAAUGCAAAGUGCAUGUGCAGCCAUCAGCUAUUAUUUUGUUUUGGUUUUUGAU--------- .((((((((((((((.(((....))).)))))).))))))))((((((((.((((.....)))).))))((((..(((.........))).)))).))))..--------- ( -33.70) >DroEre_CAF1 97729 104 + 1 UCAGUUGCUUUGAGCCGCGAACU--CCGCUCAGCGGCAAUUGCAAAUGCAAAUGCAAAGUGCAUGUGCAGCCACAAGCUAUUAUUUUGUUUUGGUUUUUCGUUUUG----- .((((((((((((((.(......--).)))))).))))))))....((((.((((.....)))).))))((((.((((.........))))))))...........----- ( -28.90) >DroYak_CAF1 96010 104 + 1 UCAGUUGCUUUGAGCCGAGAACU--CCGCUCAGCGGCAA-----AUUGCAAAUGCAAAGUGCAUGUGCAGCCAUCACCUAAUAUUUUGUUUUGGUUUUUGAUUUUUGUAUU (((((((((((((((.((....)--).)))))).)))))-----..((((.((((.....)))).))))((((..((..........))..))))..)))).......... ( -26.20) >consensus UCAGUUGUUUUGAGCCGAGAACUCUCCGCUCAGCGGCGACUGCGAAUGCAAAUGCAAAGUGCAUGUGCAGCCAUCAGCUAUUAUUUUGUUUUGGUUUUUGAU_________ .((((((((((((((.(((....))).)))))).))))))))....((((.((((.....)))).))))((((...((.........))..))))................ (-27.40 = -28.40 + 1.00)



| Location | 7,068,878 – 7,068,979 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 87.43 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -21.16 |
| Energy contribution | -21.96 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7068878 101 - 23771897 ---------AUCAAAAACCAAAACAAAAUAA-AGCUGUUGGCUGCACAUGCACUUUGCAUUUGCAUUCGCAGUCGCCGCUGAGCGGAGAGUUCUCGGCUCAAAACAACUGA ---------.(((..................-(((.((.((((((..(((((.........)))))..)))))))).)))((((.(((....))).))))........))) ( -29.50) >DroSec_CAF1 81457 102 - 1 ---------AUCAAAAACCAAAACAAAAUAAUAGCUGAUGGCUGCACAUGCACUUUGCAUUUGCAUUCGCAGUCGCCGCUGAGCGGAGAGUUCUCGGCUCAAAACAACUGA ---------.(((...................(((.(.(((((((..(((((.........)))))..)))))))).)))((((.(((....))).))))........))) ( -29.40) >DroSim_CAF1 114189 102 - 1 ---------AUCAAAAACCAAAACAAAAUAAUAGCUGAUGGCUGCACAUGCACUUUGCAUUUGCAUUCGCAGUCGCCGCUGAGCGGAGAGUUCUCGGCUCAAAACAACUGA ---------.(((...................(((.(.(((((((..(((((.........)))))..)))))))).)))((((.(((....))).))))........))) ( -29.40) >DroEre_CAF1 97729 104 - 1 -----CAAAACGAAAAACCAAAACAAAAUAAUAGCUUGUGGCUGCACAUGCACUUUGCAUUUGCAUUUGCAAUUGCCGCUGAGCGG--AGUUCGCGGCUCAAAGCAACUGA -----............................(((((..(.((((.((((.....)))).)))).)..)....(((((.((((..--.)))))))))...))))...... ( -28.90) >DroYak_CAF1 96010 104 - 1 AAUACAAAAAUCAAAAACCAAAACAAAAUAUUAGGUGAUGGCUGCACAUGCACUUUGCAUUUGCAAU-----UUGCCGCUGAGCGG--AGUUCUCGGCUCAAAGCAACUGA ..............................((((.....(((((((.((((.....)))).))))..-----..)))(((((((.(--(....)).))))..)))..)))) ( -25.90) >consensus _________AUCAAAAACCAAAACAAAAUAAUAGCUGAUGGCUGCACAUGCACUUUGCAUUUGCAUUCGCAGUCGCCGCUGAGCGGAGAGUUCUCGGCUCAAAACAACUGA .......................................(((((((.((((.....)))).)))).........)))..(((((.(((....))).))))).......... (-21.16 = -21.96 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:34 2006