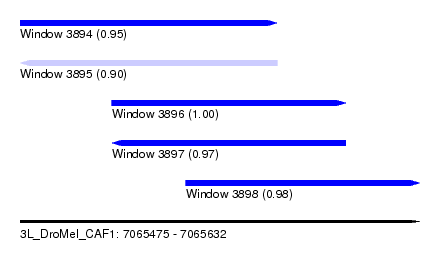
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,065,475 – 7,065,632 |
| Length | 157 |
| Max. P | 0.997829 |

| Location | 7,065,475 – 7,065,576 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.57 |
| Mean single sequence MFE | -22.92 |
| Consensus MFE | -14.80 |
| Energy contribution | -15.08 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7065475 101 + 23771897 UUAAAGCCGCCCAAGCGGCAGGCACCGGACAAUUGUAUUGGCCAAAAA-AAAAAAGA--------UUUAUAUAUGUAUAUAAAGAAAUGUACAG----AAAAAAUAUAUGCCCA .....(((((....))))).((((..((.((((...)))).)).....-........--------........(((((((......))))))).----..........)))).. ( -22.90) >DroSec_CAF1 78053 102 + 1 UUAAAGCCGCCCAAGCGGCAGGCACCGGACAAUUGUAUUGGGCCAAAAAAAAAGAGA--------UGUAUAUAUGUAUAUAAAGAUACGUACAC----AAAAAAUAAAUGCCCA .....(((((....))))).(((.((((((....)).))))))).............--------(((...(((((((......))))))).))----)............... ( -23.70) >DroSim_CAF1 110778 101 + 1 UUAAAGCCGCCCAAGCGGCAGGCACCGGACAAUUGUAUUGGGCCAAAA-AAAAGAGA--------UGUAUAUGUGUAUAUAAAGAAACGUACAG----AAAAAAUAAAUGCCCA .....(((((....))))).(((.((((((....)).)))))))....-........--------......(.(((((..........))))).----)............... ( -20.80) >DroEre_CAF1 93759 99 + 1 UUAAAGCCGCCCAAGCGGCAGGCACCGGACAAUUGUGUUG-CC-AAAA-AAUAUAUACCAG--------UAUAUGUAUAUAUCGCAGCGUACAG----AAAAAGUAAAUGCCCA .....(((((....))))).((((.((.(....).)).))-))-....-.((((((((...--------.....)))))))).(((...(((..----.....)))..)))... ( -24.50) >DroYak_CAF1 92147 112 + 1 UUAAAGCCGCCCAAGCGACGAGCACCGGACAAUUGUGUUGGCC-AAAA-AAAAAAUACGCACAUAUAUAUAUGUGUGUAUAAAGGAACGUACAGCAAAAAAAAAUAAAUGCCCA .....(((((....)))(((....((((.((((...)))).))-....-.....(((((((((((....)))))))))))...))..)))...))................... ( -22.70) >consensus UUAAAGCCGCCCAAGCGGCAGGCACCGGACAAUUGUAUUGGCC_AAAA_AAAAAAGA________UGUAUAUAUGUAUAUAAAGAAACGUACAG____AAAAAAUAAAUGCCCA .....(((((....))))).((((((((((....)).))))................................((((..(......)..))))...............)))).. (-14.80 = -15.08 + 0.28)

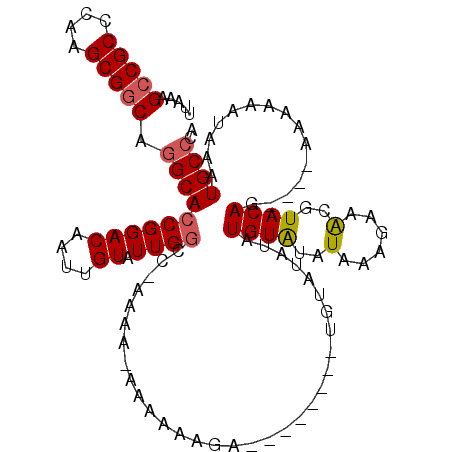

| Location | 7,065,475 – 7,065,576 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.57 |
| Mean single sequence MFE | -24.24 |
| Consensus MFE | -17.92 |
| Energy contribution | -18.08 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7065475 101 - 23771897 UGGGCAUAUAUUUUUU----CUGUACAUUUCUUUAUAUACAUAUAUAAA--------UCUUUUUU-UUUUUGGCCAAUACAAUUGUCCGGUGCCUGCCGCUUGGGCGGCUUUAA .((((((.........----...........(((((((....)))))))--------........-....(((.((((...)))).)))))))))(((((....)))))..... ( -23.00) >DroSec_CAF1 78053 102 - 1 UGGGCAUUUAUUUUUU----GUGUACGUAUCUUUAUAUACAUAUAUACA--------UCUCUUUUUUUUUGGCCCAAUACAAUUGUCCGGUGCCUGCCGCUUGGGCGGCUUUAA .(((((((.......(----(((((..((....))..))))))......--------.............((..((((...)))).)))))))))(((((....)))))..... ( -24.10) >DroSim_CAF1 110778 101 - 1 UGGGCAUUUAUUUUUU----CUGUACGUUUCUUUAUAUACACAUAUACA--------UCUCUUUU-UUUUGGCCCAAUACAAUUGUCCGGUGCCUGCCGCUUGGGCGGCUUUAA .(((((((........----.((((..(......)..))))........--------........-....((..((((...)))).)))))))))(((((....)))))..... ( -20.10) >DroEre_CAF1 93759 99 - 1 UGGGCAUUUACUUUUU----CUGUACGCUGCGAUAUAUACAUAUA--------CUGGUAUAUAUU-UUUU-GG-CAACACAAUUGUCCGGUGCCUGCCGCUUGGGCGGCUUUAA .(((((((.((...((----.(((..((((.(((((((((.....--------...)))))))))-...)-))-).))).))..))..)))))))(((((....)))))..... ( -28.10) >DroYak_CAF1 92147 112 - 1 UGGGCAUUUAUUUUUUUUUGCUGUACGUUCCUUUAUACACACAUAUAUAUAUGUGCGUAUUUUUU-UUUU-GGCCAACACAAUUGUCCGGUGCUCGUCGCUUGGGCGGCUUUAA .(((((((..........................((((.((((((....)))))).)))).....-....-((.(((.....))).)))))))))(((((....)))))..... ( -25.90) >consensus UGGGCAUUUAUUUUUU____CUGUACGUUUCUUUAUAUACAUAUAUACA________UCUCUUUU_UUUU_GGCCAAUACAAUUGUCCGGUGCCUGCCGCUUGGGCGGCUUUAA .((((((..............((((.((......)).))))..............................((.(((.....))).)).))))))(((((....)))))..... (-17.92 = -18.08 + 0.16)


| Location | 7,065,511 – 7,065,603 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 73.91 |
| Mean single sequence MFE | -24.81 |
| Consensus MFE | -15.17 |
| Energy contribution | -16.00 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.94 |
| SVM RNA-class probability | 0.997829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7065511 92 + 23771897 AUUGG--CCAAAAA-AAAAAAGA--------UUUAUAUAUGUAUAUAAAGAAAUGUACAG----AAAAAAUAUAUGCCCAAUUGCCGGACAGCCGGUUUGGGGCAUA .....--.......-........--------........(((((((......))))))).----........(((((((....(((((....)))))...))))))) ( -24.70) >DroSec_CAF1 78089 93 + 1 AUUGG--GCCAAAAAAAAAGAGA--------UGUAUAUAUGUAUAUAAAGAUACGUACAC----AAAAAAUAAAUGCCCAAUUGCCGGACAGCCGGUUUGGGGCAUA .....--................--------(((...(((((((......))))))).))----)........((((((....(((((....)))))...)))))). ( -24.00) >DroSim_CAF1 110814 92 + 1 AUUGG--GCCAAAA-AAAAGAGA--------UGUAUAUGUGUAUAUAAAGAAACGUACAG----AAAAAAUAAAUGCCCAAUUGCCGGACAGCCGGUUUGGGGCAUA .....--.......-........--------......(.(((((..........))))).----)........((((((....(((((....)))))...)))))). ( -21.10) >DroEre_CAF1 93795 90 + 1 GUUG---CC-AAAA-AAUAUAUACCAG--------UAUAUGUAUAUAUCGCAGCGUACAG----AAAAAGUAAAUGCCCAAUUGCCGGACAGCCGGUUUGGGGCAUA ((((---(.-....-.((((((((...--------.....)))))))).)))))......----.........((((((....(((((....)))))...)))))). ( -28.80) >DroYak_CAF1 92183 103 + 1 GUUGG--CC-AAAA-AAAAAAUACGCACAUAUAUAUAUGUGUGUAUAAAGGAACGUACAGCAAAAAAAAAUAAAUGCCCAAUUGCCGGACAGCCGGUUUGGGGCAUA ((((.--.(-....-.....(((((((((((....)))))))))))........)..))))............((((((....(((((....)))))...)))))). ( -34.73) >DroAna_CAF1 95076 76 + 1 GUUGCCGGC-AAAA-AGCAA------------------------AUACGGUGGCCAGCAG----AAAAAAAAGAAG-CAAAUUGCCGAACAGCCGGUUUGGAGCAUA ((..(((..-....-.....------------------------...)))..))......----...........(-(.....((((......)))).....))... ( -15.54) >consensus AUUGG__CC_AAAA_AAAAAAGA________U_UAUAUAUGUAUAUAAAGAAACGUACAG____AAAAAAUAAAUGCCCAAUUGCCGGACAGCCGGUUUGGGGCAUA .........................................................................((((((....(((((....)))))...)))))). (-15.17 = -16.00 + 0.83)


| Location | 7,065,511 – 7,065,603 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 73.91 |
| Mean single sequence MFE | -19.01 |
| Consensus MFE | -11.18 |
| Energy contribution | -11.40 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7065511 92 - 23771897 UAUGCCCCAAACCGGCUGUCCGGCAAUUGGGCAUAUAUUUUUU----CUGUACAUUUCUUUAUAUACAUAUAUAAA--------UCUUUUUU-UUUUUGG--CCAAU (((((((....((((....)))).....)))))))........----...........(((((((....)))))))--------........-.......--..... ( -17.30) >DroSec_CAF1 78089 93 - 1 UAUGCCCCAAACCGGCUGUCCGGCAAUUGGGCAUUUAUUUUUU----GUGUACGUAUCUUUAUAUACAUAUAUACA--------UCUCUUUUUUUUUGGC--CCAAU ...........((((....))))..(((((((..........(----(((((..((....))..))))))....((--------............))))--))))) ( -19.10) >DroSim_CAF1 110814 92 - 1 UAUGCCCCAAACCGGCUGUCCGGCAAUUGGGCAUUUAUUUUUU----CUGUACGUUUCUUUAUAUACACAUAUACA--------UCUCUUUU-UUUUGGC--CCAAU ...........((((....))))..(((((((...........----.((((..(......)..))))......((--------........-...))))--))))) ( -15.20) >DroEre_CAF1 93795 90 - 1 UAUGCCCCAAACCGGCUGUCCGGCAAUUGGGCAUUUACUUUUU----CUGUACGCUGCGAUAUAUACAUAUA--------CUGGUAUAUAUU-UUUU-GG---CAAC .((((((....((((....)))).....)))))).........----......((((.(((((((((.....--------...)))))))))-...)-))---)... ( -23.60) >DroYak_CAF1 92183 103 - 1 UAUGCCCCAAACCGGCUGUCCGGCAAUUGGGCAUUUAUUUUUUUUUGCUGUACGUUCCUUUAUACACACAUAUAUAUAUGUGCGUAUUUUUU-UUUU-GG--CCAAC .((((((....((((....)))).....))))))............((((...........((((.((((((....)))))).)))).....-...)-))--).... ( -23.81) >DroAna_CAF1 95076 76 - 1 UAUGCUCCAAACCGGCUGUUCGGCAAUUUG-CUUCUUUUUUUU----CUGCUGGCCACCGUAU------------------------UUGCU-UUUU-GCCGGCAAC ..((((......(((.((..(((((.....-............----.)))))..)))))...------------------------..((.-....-)).)))).. ( -15.07) >consensus UAUGCCCCAAACCGGCUGUCCGGCAAUUGGGCAUUUAUUUUUU____CUGUACGUUUCUUUAUAUACAUAUAUA_A________UCUUUUUU_UUUU_GC__CCAAC .((((((....((((....)))).....))))))......................................................................... (-11.18 = -11.40 + 0.22)
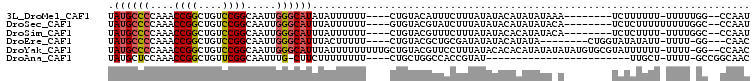
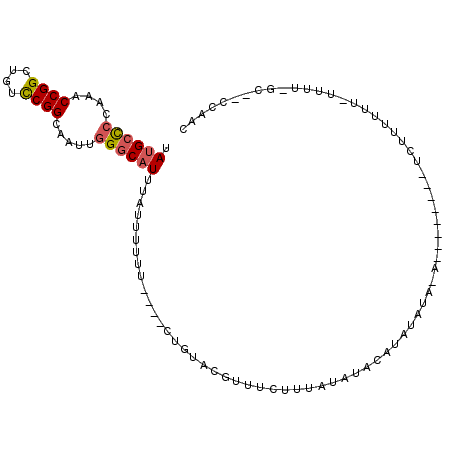
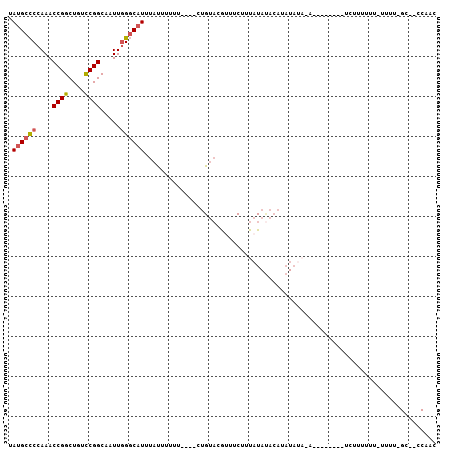
| Location | 7,065,540 – 7,065,632 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 93.48 |
| Mean single sequence MFE | -24.55 |
| Consensus MFE | -22.56 |
| Energy contribution | -22.56 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7065540 92 + 23771897 GUAUAUAAAGAAAUGUACAGAAAAAAUAUAUGCCCAAUUGCCGGACAGCCGGUUUGGGGCAUAAAAGCCGAAAGUAACACCAAAAAGGCCAG ((((((......))))))..........(((((((....(((((....)))))...)))))))...(((.................)))... ( -27.33) >DroSec_CAF1 78119 92 + 1 GUAUAUAAAGAUACGUACACAAAAAAUAAAUGCCCAAUUGCCGGACAGCCGGUUUGGGGCAUAAAAGCCGAAAGUAACACCAAAAAGGCCAG ((((......))))...............((((((....(((((....)))))...))))))....(((.................)))... ( -23.63) >DroEre_CAF1 93822 92 + 1 GUAUAUAUCGCAGCGUACAGAAAAAGUAAAUGCCCAAUUGCCGGACAGCCGGUUUGGGGCAUAAAAGCCGAAAGUAACACCAAAAAGGCCAG .........((.((.(((.......))).((((((....(((((....)))))...))))))....))(....).............))... ( -22.70) >consensus GUAUAUAAAGAAACGUACAGAAAAAAUAAAUGCCCAAUUGCCGGACAGCCGGUUUGGGGCAUAAAAGCCGAAAGUAACACCAAAAAGGCCAG ((((..........))))...........((((((....(((((....)))))...))))))....(((.................)))... (-22.56 = -22.56 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:29 2006