
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,046,366 – 7,046,478 |
| Length | 112 |
| Max. P | 0.878363 |

| Location | 7,046,366 – 7,046,478 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.36 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -15.93 |
| Energy contribution | -17.57 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7046366 112 - 23771897 UCGCGAUGCUGUUUCGCUGAUGUUUCUCCGGCUGAUUGCGUUGACGAAAUCGCAUUGACGGUC---GAAAACACAGCGAAAAAUAGUAAUAA--ACCAUUUUAAUAAAA-UAUUCAAA ......(((((((((((((.(((((...((((((..((((((.....)).))))....)))))---).)))))))))))..)))))))....--...............-........ ( -29.10) >DroSec_CAF1 59073 113 - 1 UCGCGAUGCUGUUUCGCUGAUGUUUCUCUGGCUGAUUGCGUUGACGAAAUCGCAUUGACAGUC---GAAAACACAGCGAAAAAUAAUAAAAA--ACCAUUUAAAUAAAAAAAUCCAAA ...........((((((((.(((((.((.(((((..((((((.....)).))))....)))))---)))))))))))))))...........--........................ ( -25.50) >DroSim_CAF1 91721 112 - 1 UCGCGAUGCUGUUUCGCUGAUGUUUCUCCGGCUGAUUGCGUUGACGAAAUCGCAUUGACGGUC---GAAAACACAGCGAAAAAUAAUAAUAA--ACCAUUUAAAUAAAA-AAUUCAAA ...........((((((((.(((((...((((((..((((((.....)).))))....)))))---).)))))))))))))...........--...............-........ ( -25.70) >DroEre_CAF1 68085 112 - 1 UCGCGAUGCUGUUUCGCUGAUGUUUCUCCGGCUGAUUGCGUUGACGAAAUCGCAUUGACGGUC---GACAACACAGCGAAAAAUAAUAAUGC--AUCAUUGUAUUAAAA-AAUUCACA ....(((((..((((((((.((((....((((((..((((((.....)).))))....)))))---)..)))))))))))).........))--)))............-........ ( -30.50) >DroWil_CAF1 102738 97 - 1 --------------CGCUG-UGCUUGUCUCGAUGAUUGCCUUGACAAAAUCACAUUGACAGCUCAGAAAAACACAGAGAAAACACAGAGAAAACACCACAGCAACAAAC-AGU----- --------------.((((-((....(((((.(((((..........))))))........(((...........)))........))))......)))))).......-...----- ( -17.50) >DroYak_CAF1 67407 112 - 1 UCGCGAUGCUGUUUCGCUGAUGUUUCUCCGGCUGAUUGCGUUGAUGAAAUCGCAUUGACGCUC---GAACACACAGCGAAAAAUAAUAAUAA--ACCAUUUUAUUAAAA-AAAACGCA ..(((......((((((((.((((((((.....))..((((..(((......)))..))))..---))).))))))))))).....((((((--(....)))))))...-....))). ( -30.10) >consensus UCGCGAUGCUGUUUCGCUGAUGUUUCUCCGGCUGAUUGCGUUGACGAAAUCGCAUUGACGGUC___GAAAACACAGCGAAAAAUAAUAAUAA__ACCAUUUUAAUAAAA_AAUUCAAA ...........((((((((.(((((....(((((..((((.(......).))))....))))).....)))))))))))))..................................... (-15.93 = -17.57 + 1.64)

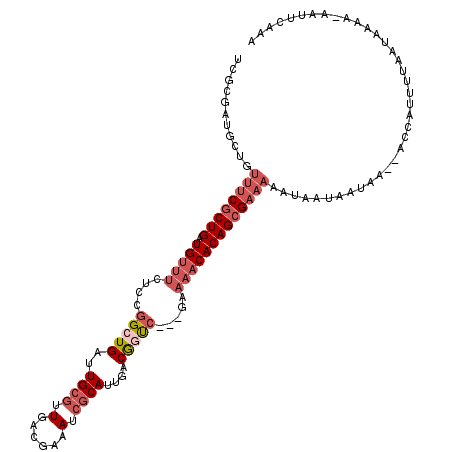

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:19 2006