| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,043,100 – 7,043,199 |
| Length | 99 |
| Max. P | 0.935653 |

| Location | 7,043,100 – 7,043,199 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
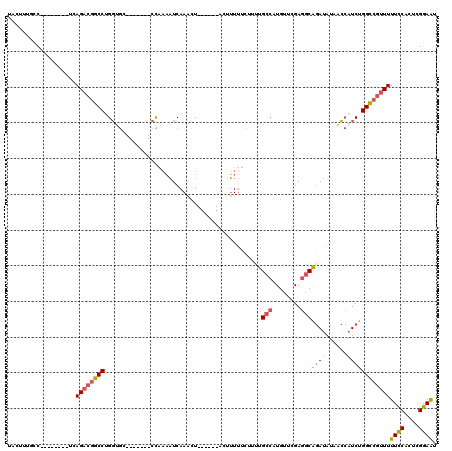
| Mean pairwise identity | 73.58 |
| Mean single sequence MFE | -26.17 |
| Consensus MFE | -16.38 |
| Energy contribution | -16.85 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.71 |
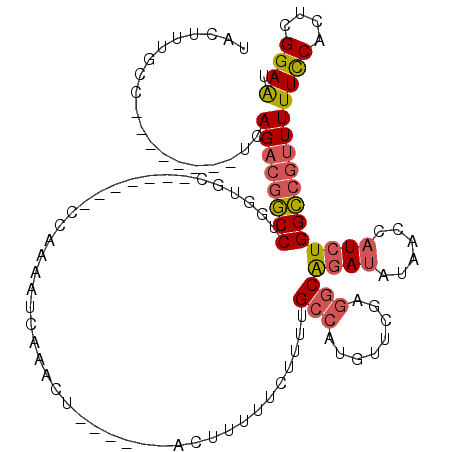
| Structure conservation index | 0.63 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
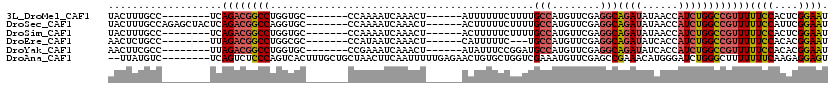
>3L_DroMel_CAF1 7043100 99 - 23771897 UACUUUGCC--------UCAGACGGCCUGGUGC-------CCAAAAUCAAACU------AUUUUUUCUUUUGCCAUGUUCGAGGCAGAUAUAACCAUCUGGCCGUUUUUCCACUCGGAAU .........--------..((((((((.((((.-------..(((((......------)))))....((((((........))))))......)))).))))))))((((....)))). ( -25.10) >DroSec_CAF1 55894 107 - 1 UACUUUGCCAGAGCUACUCAGACGGCCAGGUGC-------CCAAAAUCAAACU------ACUUUUUCUUUUGCCAUGUUCGAGGCAGAUAUAACCAUCUGGCCGUUUUUCCAUUCGGAAU ..........(((...)))(((((((((((((.-------..((((.......------...))))..((((((........))))))......)))))))))))))((((....)))). ( -29.30) >DroSim_CAF1 88462 99 - 1 UACUUUGCC--------UCAGACGGCCUGGUGC-------CCAAAAUCAAACU------ACUUUUUCUUUUGCCAUGUUCGAGGCAGAUAUAACCAUCUGGCCGUUUUUCCACUCGGAAU .........--------..((((((((.((((.-------..((((.......------...))))..((((((........))))))......)))).))))))))((((....)))). ( -24.00) >DroEre_CAF1 64720 96 - 1 AACUCUGCC--------UUAGACGGCCUGGCGC-------CCAUAAUCAAACU------CAUUUUUC---UGCCAUGUUCGAGGCAGAUAUCACCAUCUGGCCGUUUUUCCACACGGAAU .........--------..(((((((((((...-------)))..........------......((---((((........))))))...........))))))))((((....)))). ( -25.10) >DroYak_CAF1 63977 99 - 1 AACUUCGCC--------UUAGACGGCCUGGUGC-------CCGAAAUCAAACU------AUAUUUCCGGAUGCCAUGUUCGAGGCAGAUAUCACCAUCUGGCCGUUUUUCCACACGGAAU .........--------..((((((((((((((-------(.(((((......------..))))).)).((((........)))).....)))))...))))))))((((....)))). ( -30.20) >DroAna_CAF1 72668 110 - 1 --UUAUGUC--------UCAGUCUCCCAGUCACUUUGCUGCUAACUUCAAUUUUUGAGAACUGUGCUGGUCGAAAUGUUCGAGCCGAAACAUGGGAUCUGGGCUUUUUUUCAAGAGGAGU --......(--------((((..(((((.((.....((.((....((((.....))))....)))).((((((.....)))).))))....))))).)))))((((((.....)))))). ( -23.30) >consensus UACUUUGCC________UCAGACGGCCUGGUGC_______CCAAAAUCAAACU______ACUUUUUCUUUUGCCAUGUUCGAGGCAGAUAUAACCAUCUGGCCGUUUUUCCACUCGGAAU ...................((((((((............................................(((........)))((((......))))))))))))((((....)))). (-16.38 = -16.85 + 0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:17 2006