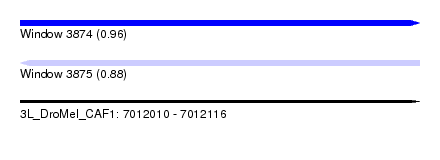
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,012,010 – 7,012,116 |
| Length | 106 |
| Max. P | 0.964764 |

| Location | 7,012,010 – 7,012,116 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.58 |
| Mean single sequence MFE | -54.35 |
| Consensus MFE | -42.40 |
| Energy contribution | -42.52 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
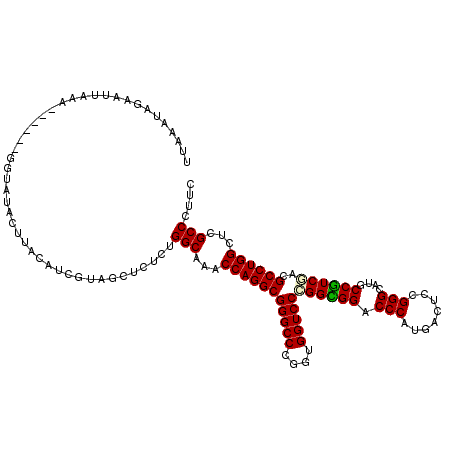
>3L_DroMel_CAF1 7012010 106 + 23771897 GAAGGGCGAGCCAGGCGUCGACGGGCUGCCCGGAGUCAUGGGUCCGCCGGGACCACCGGGCCCGCCUGGUUUGCCCGAGAAUUACGAUGUAAGUAUUCCA--------------AUGGAA ...((((((((((((((.(..((((...))))..).)..(((((((..(....)..))))))))))))))))))))(.((((.((.......))))))).--------------...... ( -52.70) >DroVir_CAF1 51896 114 + 1 GAAGGGCGAGCCAGGCGUCGACGGCAUGCCCGGAGUGAUGGGUCCGCCCGGACCUCCGGGCCCGCCUGGCUUGCCGGAGAGCUAUGAUGUGAGUACAAC------UUUAAUUCUAUUCAA ....((((((((((((((.....))..((((((((...((((....))))...))))))))..)))))))))))).........(((...((((.....------....))))...))). ( -55.10) >DroGri_CAF1 45763 114 + 1 GAAGGGCGAGCCAGGCGUUGACGGCAUGCCCGGCGUCAUGGGUCCACCAGGACCUGCGGGCCCGCCUGGCUUGCCAGAGAGCUACGAUGUAAGUACACC------UUUAAUUCGAUUUAC (((.((((((((((((..(((((.(......).))))).((((((..(((...))).))))))))))))))))))((((...(((.......)))...)------)))..)))....... ( -50.30) >DroMoj_CAF1 45307 114 + 1 GAAGGGCGAGCCAGGCGUCGACGGCAUGCCCGGAGUGAUGGGUCCACCCGGACCCCCGGGCCCGCCUGGCUUGCCAGAGAGCUAUGAUGUGAGUACAAG------UUGAAUUCGUUUUAA ....((((((((((((((.....))..((((((......((((((....))))))))))))..)))))))))))).........(((..(((((.(...------..).)))))..))). ( -54.00) >DroAna_CAF1 43393 120 + 1 GAAGGGCGAGCCAGGCGUCGACGGUAUGCCCGGGGUCGUGGGUCCGCCGGGACCACCGGGCCCGCCUGGUUUGCCCGAGAAUUACGAUGUAAGUAUUCCGAAUAUUUUAAUUCUAUUUAA ...((((((((((((((....)((...((((((((((.(((.....))).)))).))))))))))))))))))))).(((((((.(((((..(.....)..))))).)))))))...... ( -61.10) >DroPer_CAF1 46605 119 + 1 GAAGGGCGAGCCAGGCGUCGAUGGUCUGCCCGGAGUCGCGGGUCCACCGGGACCAGCAGGCCCGCCUGGUUUGCCAGAGAACUACGAUGUAAGUAUCCUUCAAGAUAU-AACCUAUUUAA (((((((((((((((((.....(((((((...(.(((.(((.....))).)))).))))))))))))))))))))..........((((....)))))))).......-........... ( -52.90) >consensus GAAGGGCGAGCCAGGCGUCGACGGCAUGCCCGGAGUCAUGGGUCCACCGGGACCACCGGGCCCGCCUGGCUUGCCAGAGAACUACGAUGUAAGUACACC______UUUAAUUCUAUUUAA ....(((((((((((((....)((...((((((.......(((((....))))).))))))))))))))))))))............................................. (-42.40 = -42.52 + 0.11)



| Location | 7,012,010 – 7,012,116 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.58 |
| Mean single sequence MFE | -44.93 |
| Consensus MFE | -35.10 |
| Energy contribution | -34.88 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7012010 106 - 23771897 UUCCAU--------------UGGAAUACUUACAUCGUAAUUCUCGGGCAAACCAGGCGGGCCCGGUGGUCCCGGCGGACCCAUGACUCCGGGCAGCCCGUCGACGCCUGGCUCGCCCUUC ......--------------.(((((((.......)).))))).((((...(((((((((((....)))))((((((.(((........)))....))))))..))))))...))))... ( -42.20) >DroVir_CAF1 51896 114 - 1 UUGAAUAGAAUUAAA------GUUGUACUCACAUCAUAGCUCUCCGGCAAGCCAGGCGGGCCCGGAGGUCCGGGCGGACCCAUCACUCCGGGCAUGCCGUCGACGCCUGGCUCGCCCUUC .(((.((.(((....------))).)).)))..............(((.(((((((((.((((((((....(((....)))....))))))))..........))))))))).))).... ( -48.50) >DroGri_CAF1 45763 114 - 1 GUAAAUCGAAUUAAA------GGUGUACUUACAUCGUAGCUCUCUGGCAAGCCAGGCGGGCCCGCAGGUCCUGGUGGACCCAUGACGCCGGGCAUGCCGUCAACGCCUGGCUCGCCCUUC ...............------(((((....)))))..........(((.((((((((((((..(((.(.(((((((.........)))))))).))).)))..))))))))).))).... ( -45.20) >DroMoj_CAF1 45307 114 - 1 UUAAAACGAAUUCAA------CUUGUACUCACAUCAUAGCUCUCUGGCAAGCCAGGCGGGCCCGGGGGUCCGGGUGGACCCAUCACUCCGGGCAUGCCGUCGACGCCUGGCUCGCCCUUC ...............------........................(((.(((((((((.((((((((((((....))))))......))))))..........))))))))).))).... ( -46.40) >DroAna_CAF1 43393 120 - 1 UUAAAUAGAAUUAAAAUAUUCGGAAUACUUACAUCGUAAUUCUCGGGCAAACCAGGCGGGCCCGGUGGUCCCGGCGGACCCACGACCCCGGGCAUACCGUCGACGCCUGGCUCGCCCUUC .......((((......))))(((.(((.......))).)))..((((...(((((((....(((((..(((((.((........)))))))..)))))....)))))))...))))... ( -46.00) >DroPer_CAF1 46605 119 - 1 UUAAAUAGGUU-AUAUCUUGAAGGAUACUUACAUCGUAGUUCUCUGGCAAACCAGGCGGGCCUGCUGGUCCCGGUGGACCCGCGACUCCGGGCAGACCAUCGACGCCUGGCUCGCCCUUC ......(((..-.(((..(((((....))).))..)))..)))..(((...(((((((((((....)))))((((((.((((......))))....))))))..))))))...))).... ( -41.30) >consensus UUAAAUAGAAUUAAA______GGUAUACUUACAUCGUAGCUCUCUGGCAAACCAGGCGGGCCCGGUGGUCCCGGCGGACCCAUGACUCCGGGCAUGCCGUCGACGCCUGGCUCGCCCUUC .............................................(((...(((((((((((....)))))((((((.(((........)))....))))))..))))))...))).... (-35.10 = -34.88 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:08 2006