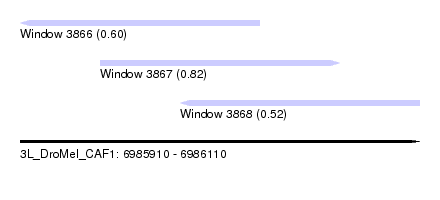
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,985,910 – 6,986,110 |
| Length | 200 |
| Max. P | 0.820312 |

| Location | 6,985,910 – 6,986,030 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -30.83 |
| Energy contribution | -30.83 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6985910 120 - 23771897 GGUAAUCAAUGCACAAAUGUGGCCAGUUUGGGAUUGCAUUCGCAAUUCCGCAUAUAUGCAUACAAUUCCCCUCCACUUUUGCGGACACUUGUGCGUGGAAAUAAACCAACCGCAGACACU (((.....((((((((..(((.((.....((((((((....))))))))(((....))).......................)).)))))))))))((.......)).)))......... ( -32.00) >DroSec_CAF1 55749 120 - 1 GGUAAUCAAUGCACAAAUGUGGCCAGUUUGGGAUUGCAUUCGCAAUUCCGCAUAUAUGUAUACAAUUCCCCUCCACUUUUGCGGACACUUGUGCGUGGAAAUAAACCAACCGCAGACACU (((.....((((((((..(((.((.....((((((((....))))))))(((...........................))))).)))))))))))((.......)).)))......... ( -30.83) >DroSim_CAF1 55546 120 - 1 GGUAAUCAAUGCACAAAUGUGGCCAGUUUGGGAUUGCAUUCGCAAUUCCGCAUAUAUGUAUACAAUUCCCCUCCACUUUUGCGGACACUUGUGCGUGGAAAUAAACCAACCGCAGACACU (((.....((((((((..(((.((.....((((((((....))))))))(((...........................))))).)))))))))))((.......)).)))......... ( -30.83) >consensus GGUAAUCAAUGCACAAAUGUGGCCAGUUUGGGAUUGCAUUCGCAAUUCCGCAUAUAUGUAUACAAUUCCCCUCCACUUUUGCGGACACUUGUGCGUGGAAAUAAACCAACCGCAGACACU (((.....((((((((..(((.((.....((((((((....))))))))(((...........................))))).)))))))))))((.......)).)))......... (-30.83 = -30.83 + 0.00)

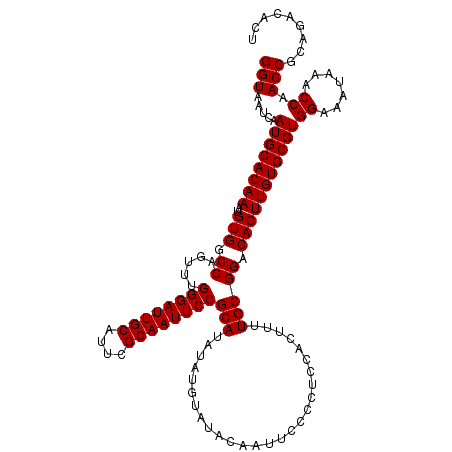
| Location | 6,985,950 – 6,986,070 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -31.46 |
| Energy contribution | -30.80 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6985950 120 + 23771897 AAAAGUGGAGGGGAAUUGUAUGCAUAUAUGCGGAAUUGCGAAUGCAAUCCCAAACUGGCCACAUUUGUGCAUUGAUUACCCAGCGAAACGGAAGUGCCGCUUUAAUCUCAUAAUUUUACA .(((((.(((((((((((.(((((((.(((.((.(((((....))))).((.....)))).))).)))))))))))).)))((((..((....))..)))).....)))...)))))... ( -36.90) >DroSec_CAF1 55789 120 + 1 AAAAGUGGAGGGGAAUUGUAUACAUAUAUGCGGAAUUGCGAAUGCAAUCCCAAACUGGCCACAUUUGUGCAUUGAUUACCCAGCGAAACGGAAGUGCCGCUUUAAUUUCAUAGUUUUACA ....((((..((.....(((((....)))))((.(((((....))))).))...))..))))..(((((....(((((...((((..((....))..)))).))))).)))))....... ( -29.50) >DroSim_CAF1 55586 120 + 1 AAAAGUGGAGGGGAAUUGUAUACAUAUAUGCGGAAUUGCGAAUGCAAUCCCAAACUGGCCACAUUUGUGCAUUGAUUACCCAGCGAAACGGAAGUGCCGCUUUAAUCUCAUAAUUUUACA ....((((..((.....(((((....)))))((.(((((....))))).))...))..))))..(((((....(((((...((((..((....))..)))).))))).)))))....... ( -31.60) >consensus AAAAGUGGAGGGGAAUUGUAUACAUAUAUGCGGAAUUGCGAAUGCAAUCCCAAACUGGCCACAUUUGUGCAUUGAUUACCCAGCGAAACGGAAGUGCCGCUUUAAUCUCAUAAUUUUACA ....((((..((.....(((((....)))))((.(((((....))))).))...))..))))..(((((....(((((...((((..((....))..)))).))))).)))))....... (-31.46 = -30.80 + -0.66)

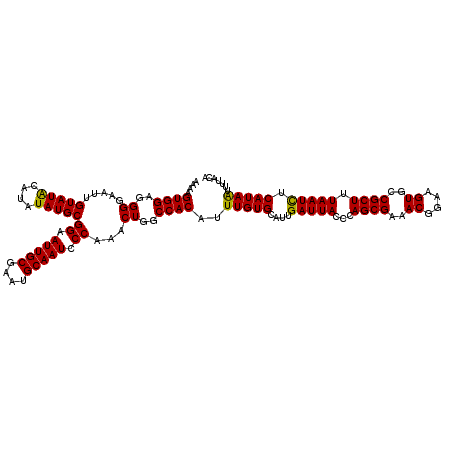
| Location | 6,985,990 – 6,986,110 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.21 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -27.77 |
| Energy contribution | -28.10 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.98 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6985990 120 - 23771897 AAAAAAGCAAUAAAAACAUAAGCUGCCAUCCGUUUGAAUCUGUAAAAUUAUGAGAUUAAAGCGGCACUUCCGUUUCGCUGGGUAAUCAAUGCACAAAUGUGGCCAGUUUGGGAUUGCAUU ......(((((.......((((((((((..((((((..((...........))(((((.(((((.((....)).)))))...)))))......))))))))).)))))))..)))))... ( -29.70) >DroSec_CAF1 55829 118 - 1 A--AAAGCAAUAAAAACAUAAGCUGCCAUCCGUUUGAAUCUGUAAAACUAUGAAAUUAAAGCGGCACUUCCGUUUCGCUGGGUAAUCAAUGCACAAAUGUGGCCAGUUUGGGAUUGCAUU .--...(((((.......((((((((((..((((((.......................(((((.((....)).)))))..(((.....))).))))))))).)))))))..)))))... ( -26.00) >DroSim_CAF1 55626 117 - 1 ---AAAGCAAUAAAAACAUAAGCUGCCAUCCGUUUGAAUCUGUAAAAUUAUGAGAUUAAAGCGGCACUUCCGUUUCGCUGGGUAAUCAAUGCACAAAUGUGGCCAGUUUGGGAUUGCAUU ---...(((((.......((((((((((..((((((..((...........))(((((.(((((.((....)).)))))...)))))......))))))))).)))))))..)))))... ( -29.70) >consensus A__AAAGCAAUAAAAACAUAAGCUGCCAUCCGUUUGAAUCUGUAAAAUUAUGAGAUUAAAGCGGCACUUCCGUUUCGCUGGGUAAUCAAUGCACAAAUGUGGCCAGUUUGGGAUUGCAUU ......(((((.......((((((((((..((((((..((...........))(((((.(((((.((....)).)))))...)))))......))))))))).)))))))..)))))... (-27.77 = -28.10 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:01 2006