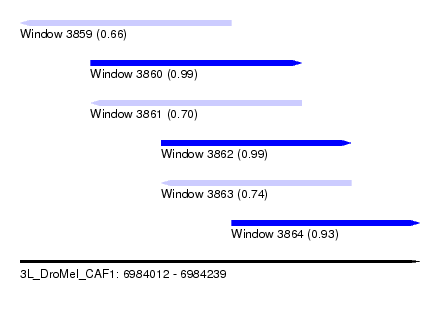
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,984,012 – 6,984,239 |
| Length | 227 |
| Max. P | 0.986777 |

| Location | 6,984,012 – 6,984,132 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -28.81 |
| Energy contribution | -28.37 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6984012 120 - 23771897 AAUGACAUCAUUAGGCGCCAAAAACAAACAGCCAUACAAAUUGAACAAUCCCCCCUAAAAGGGGAAUGUGAUUUUCAAGCAUUAUCAUACGAAUUCCUGGCGAUUUUGGUGGCGUGUCUU ((((....))))..((((((....((((..((((..............(((((.......))))).((((((...........))))))........))))...)))).))))))..... ( -30.20) >DroSec_CAF1 53831 118 - 1 AAUGGCAUCAUUAGGCGCCAAAAACAAACAGCCAUACAAAUUGAACAAUCCCCCC--AAAGGGGAAUGUGAUUUUCAAGCAUUAUCAUACGAAUUCCUGGCGAUUCUGGUGGCGUGUCUU ...(((((((((((.(((((..........((.....(((((..(((.(((((..--...))))).))))))))....))....((....)).....)))))...))))))..))))).. ( -29.20) >DroSim_CAF1 53601 118 - 1 AAUGGCAUCAUUAGGCGCCAAAAACAAACAGCCAUACAAAUUGAACAAUCCCCCC--AAAGGGGAAUGUGAUUUUCAAGCAUUAUCAUACGAAUUCCUGGCGAUUCUGGUGGCGUGUCUU ...(((((((((((.(((((..........((.....(((((..(((.(((((..--...))))).))))))))....))....((....)).....)))))...))))))..))))).. ( -29.20) >consensus AAUGGCAUCAUUAGGCGCCAAAAACAAACAGCCAUACAAAUUGAACAAUCCCCCC__AAAGGGGAAUGUGAUUUUCAAGCAUUAUCAUACGAAUUCCUGGCGAUUCUGGUGGCGUGUCUU ...(((((((((((.(((((..........((.....(((((..(((.(((((.......))))).))))))))....))....((....)).....)))))...))))))..))))).. (-28.81 = -28.37 + -0.44)

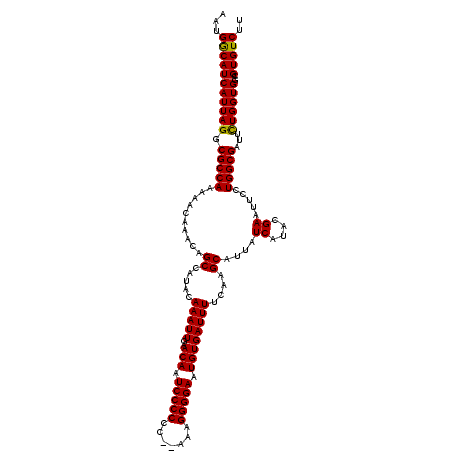
| Location | 6,984,052 – 6,984,172 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -42.77 |
| Consensus MFE | -40.06 |
| Energy contribution | -40.07 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6984052 120 + 23771897 GCUUGAAAAUCACAUUCCCCUUUUAGGGGGGAUUGUUCAAUUUGUAUGGCUGUUUGUUUUUGGCGCCUAAUGAUGUCAUUAAGUGGCUGAAGCAGAUUGCCGAACAUGCUAUUUUGAGUC (((..(((...(((.((((((.....)))))).)))......(((.((((.(((((((((.(.(((.(((((....))))).))).).))))))))).)))).))).....)))..))). ( -43.60) >DroSec_CAF1 53871 118 + 1 GCUUGAAAAUCACAUUCCCCUUU--GGGGGGAUUGUUCAAUUUGUAUGGCUGUUUGUUUUUGGCGCCUAAUGAUGCCAUUAAGUGGCCGGGGCAGAUUGCCGAACAUGCUGUUUGGAGUC ...........(((.(((((...--..))))).))).((((((((.((((..((((....(((((.(....).))))).))))..))))..))))))))((((((.....)))))).... ( -43.20) >DroSim_CAF1 53641 118 + 1 GCUUGAAAAUCACAUUCCCCUUU--GGGGGGAUUGUUCAAUUUGUAUGGCUGUUUGUUUUUGGCGCCUAAUGAUGCCAUUAAGUGGCCGCAGCAGAUUGCCGAACAUGCUGUUUGGAGUC ...........(((.(((((...--..))))).))).((((((((.((((..((((....(((((.(....).))))).))))..))))..))))))))((((((.....)))))).... ( -41.50) >consensus GCUUGAAAAUCACAUUCCCCUUU__GGGGGGAUUGUUCAAUUUGUAUGGCUGUUUGUUUUUGGCGCCUAAUGAUGCCAUUAAGUGGCCGAAGCAGAUUGCCGAACAUGCUGUUUGGAGUC ((((.(((...(((.((((((.....)))))).)))......(((.((((.(((((((((.(.(((.(((((....))))).))).).))))))))).)))).))).....))).)))). (-40.06 = -40.07 + 0.00)

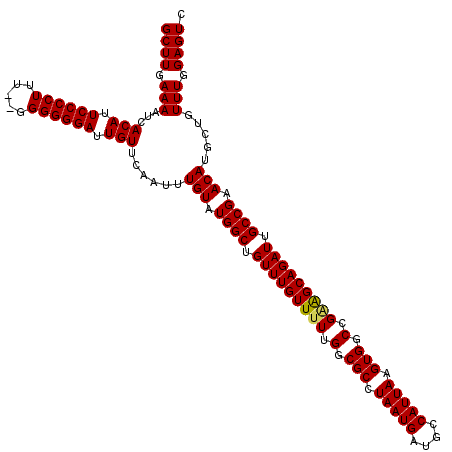
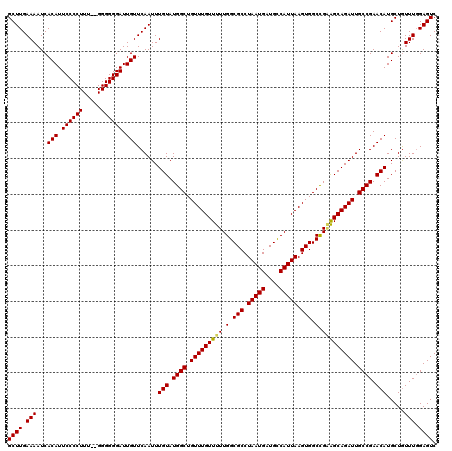
| Location | 6,984,052 – 6,984,172 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -25.45 |
| Energy contribution | -25.57 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6984052 120 - 23771897 GACUCAAAAUAGCAUGUUCGGCAAUCUGCUUCAGCCACUUAAUGACAUCAUUAGGCGCCAAAAACAAACAGCCAUACAAAUUGAACAAUCCCCCCUAAAAGGGGAAUGUGAUUUUCAAGC .....(((((.(((((((((((...........)))...(((((....)))))(((..............))).........))))..(((((.......))))))))).)))))..... ( -25.44) >DroSec_CAF1 53871 118 - 1 GACUCCAAACAGCAUGUUCGGCAAUCUGCCCCGGCCACUUAAUGGCAUCAUUAGGCGCCAAAAACAAACAGCCAUACAAAUUGAACAAUCCCCCC--AAAGGGGAAUGUGAUUUUCAAGC ...........(((((((((((.....)))..(((..(((((((....))))))).))).......................))))..(((((..--...)))))))))........... ( -28.90) >DroSim_CAF1 53641 118 - 1 GACUCCAAACAGCAUGUUCGGCAAUCUGCUGCGGCCACUUAAUGGCAUCAUUAGGCGCCAAAAACAAACAGCCAUACAAAUUGAACAAUCCCCCC--AAAGGGGAAUGUGAUUUUCAAGC ...........((.((((((((.....)))).(((..(((((((....))))))).))).......))))))........(((((((.(((((..--...))))).))).....)))).. ( -28.30) >consensus GACUCCAAACAGCAUGUUCGGCAAUCUGCUCCGGCCACUUAAUGGCAUCAUUAGGCGCCAAAAACAAACAGCCAUACAAAUUGAACAAUCCCCCC__AAAGGGGAAUGUGAUUUUCAAGC ...........(((((((((((.....)))..(((..(((((((....))))))).))).......................))))..(((((.......)))))))))........... (-25.45 = -25.57 + 0.11)

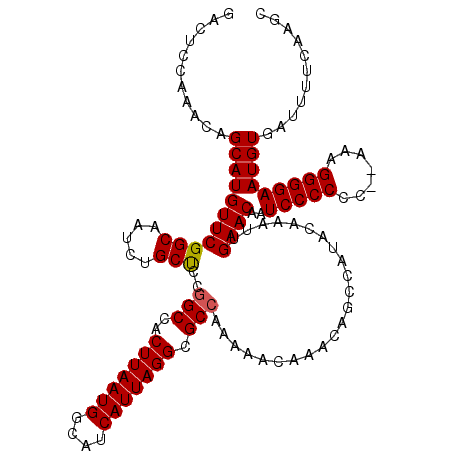
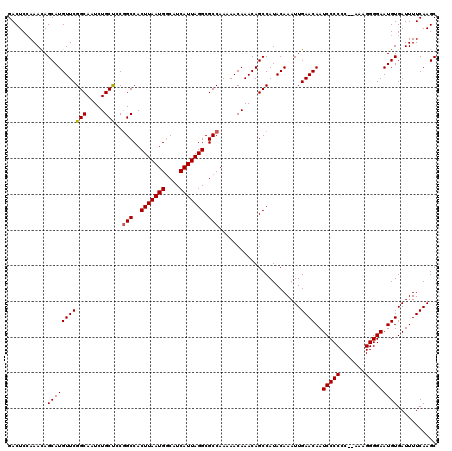
| Location | 6,984,092 – 6,984,200 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.69 |
| Mean single sequence MFE | -38.73 |
| Consensus MFE | -35.42 |
| Energy contribution | -35.53 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6984092 108 + 23771897 UUUGUAUGGCUGUUUGUUUUUGGCGCCUAAUGAUGUCAUUAAGUGGCUGAAGCAGAUUGCCGAACAUGCUAUUUUGAGUCGAAAGACUUUUGGCCAAAUAUGAUGUAA------------ ..(((.((((.(((((((((.(.(((.(((((....))))).))).).))))))))).)))).))).((((....(((((....))))).))))..............------------ ( -37.90) >DroSec_CAF1 53909 119 + 1 UUUGUAUGGCUGUUUGUUUUUGGCGCCUAAUGAUGCCAUUAAGUGGCCGGGGCAGAUUGCCGAACAUGCUGUUUGGAGUCGAAAGACUUUUGGCCAAAUAUGAUGUAA-UGCACAUAUAU ..((((((((.((((((((..(.(((.(((((....))))).))).)..)))))))).)))..((((..(((((((((((....)))).....)))))))..)))).)-))))....... ( -39.80) >DroSim_CAF1 53679 120 + 1 UUUGUAUGGCUGUUUGUUUUUGGCGCCUAAUGAUGCCAUUAAGUGGCCGCAGCAGAUUGCCGAACAUGCUGUUUGGAGUCGAAAGACUUUUGGCCAAAUAUGAUGUAAGUGCACAUAUAU ..((((((((.(((((((..((((((.(((((....))))).)).)))).))))))).)))..((((..(((((((((((....)))).....)))))))..))))..)))))....... ( -38.50) >consensus UUUGUAUGGCUGUUUGUUUUUGGCGCCUAAUGAUGCCAUUAAGUGGCCGAAGCAGAUUGCCGAACAUGCUGUUUGGAGUCGAAAGACUUUUGGCCAAAUAUGAUGUAA_UGCACAUAUAU ......((((.(((((((((.(.(((.(((((....))))).))).).))))))))).)))).((((..(((((((((((....)))).....)))))))..)))).............. (-35.42 = -35.53 + 0.12)

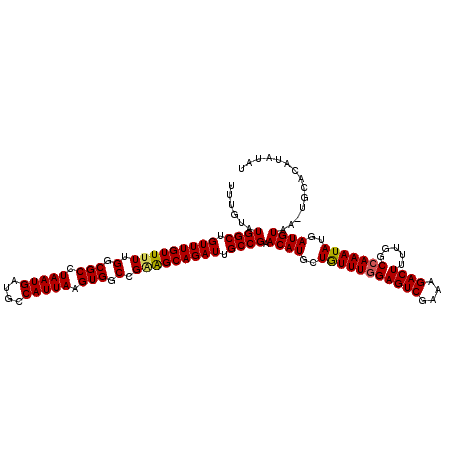
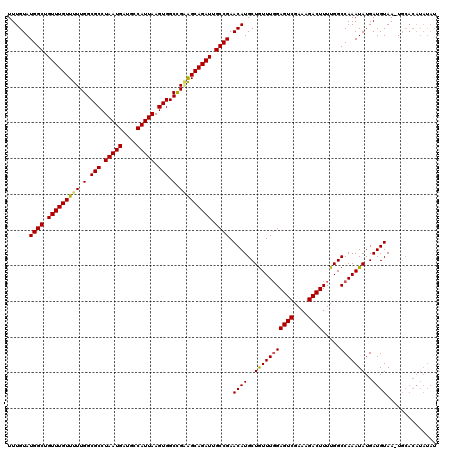
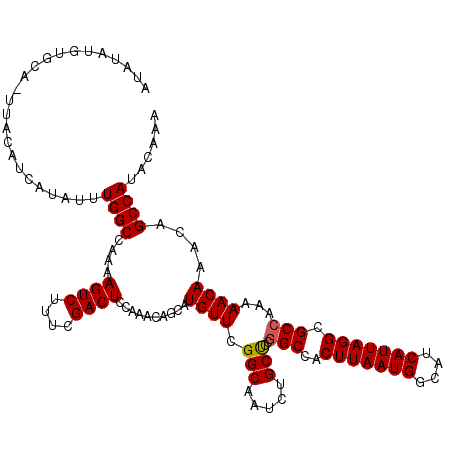
| Location | 6,984,092 – 6,984,200 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.69 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -20.05 |
| Energy contribution | -20.17 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6984092 108 - 23771897 ------------UUACAUCAUAUUUGGCCAAAAGUCUUUCGACUCAAAAUAGCAUGUUCGGCAAUCUGCUUCAGCCACUUAAUGACAUCAUUAGGCGCCAAAAACAAACAGCCAUACAAA ------------............((((....((((....))))..........((((.(((.....((....))..(((((((....))))))).)))...))))....))))...... ( -20.20) >DroSec_CAF1 53909 119 - 1 AUAUAUGUGCA-UUACAUCAUAUUUGGCCAAAAGUCUUUCGACUCCAAACAGCAUGUUCGGCAAUCUGCCCCGGCCACUUAAUGGCAUCAUUAGGCGCCAAAAACAAACAGCCAUACAAA ...((((.((.-........((..(((((...((((....))))...............(((.....)))..)))))..)).((((..(....)..))))..........)))))).... ( -27.30) >DroSim_CAF1 53679 120 - 1 AUAUAUGUGCACUUACAUCAUAUUUGGCCAAAAGUCUUUCGACUCCAAACAGCAUGUUCGGCAAUCUGCUGCGGCCACUUAAUGGCAUCAUUAGGCGCCAAAAACAAACAGCCAUACAAA ...((((.((..........((..(((((...((((....)))).....(((((.(((....))).))))).)))))..)).((((..(....)..))))..........)))))).... ( -29.00) >consensus AUAUAUGUGCA_UUACAUCAUAUUUGGCCAAAAGUCUUUCGACUCCAAACAGCAUGUUCGGCAAUCUGCUCCGGCCACUUAAUGGCAUCAUUAGGCGCCAAAAACAAACAGCCAUACAAA ........................((((....((((....))))..........((((.(((.....)))..(((..(((((((....))))))).)))...))))....))))...... (-20.05 = -20.17 + 0.11)

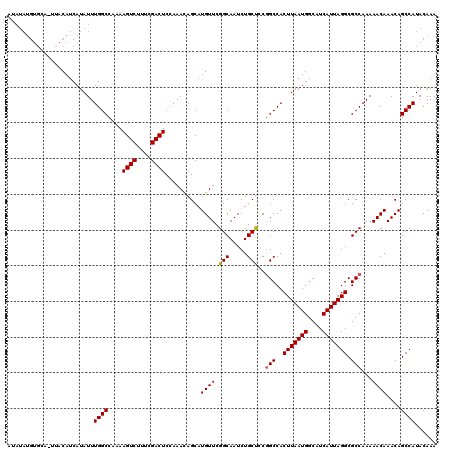
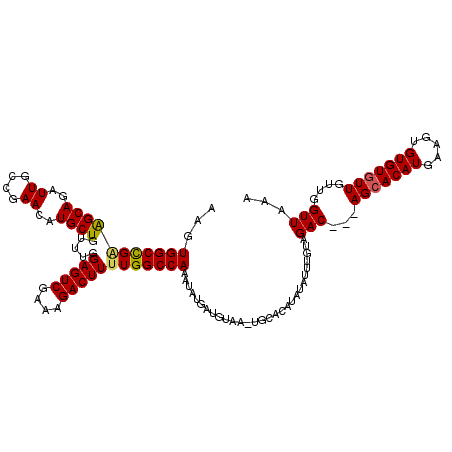
| Location | 6,984,132 – 6,984,239 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.24 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -25.15 |
| Energy contribution | -25.27 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6984132 107 + 23771897 AAGUGGCUGAAGCAGAUUGCCGAACAUGCUAUUUUGAGUCGAAAGACUUUUGGCCAAAUAUGAUGUAA-------------UGUAGACAGCAGCACAUGAAGUGUGUGUUGUUGGUUUAA ...((((..(((((..((....))..)))).....(((((....))))))..))))............-------------..(((((((((((((((.....)))))))))).))))). ( -34.30) >DroSec_CAF1 53949 116 + 1 AAGUGGCCGGGGCAGAUUGCCGAACAUGCUGUUUGGAGUCGAAAGACUUUUGGCCAAAUAUGAUGUAA-UGCACAUAUAUUUGUAGAC---AGCACAUGAAGUGUGUGUUGUUGGUUAAA ...(((((.((.((((...((((((.....))))))((((....)))))))).))(((((((.(((..-...))))))))))...(((---(((((((.....))))))))))))))).. ( -39.70) >DroSim_CAF1 53719 117 + 1 AAGUGGCCGCAGCAGAUUGCCGAACAUGCUGUUUGGAGUCGAAAGACUUUUGGCCAAAUAUGAUGUAAGUGCACAUAUAUUUGUAGAC---AGAACAUGAAGUGUGUGUUGCUGGUUAAA ...(((((((((((.((...(...((((((((((((((((....))))).....((((((((.(((......))))))))))))))))---))..))))..).)).)))))).))))).. ( -36.40) >consensus AAGUGGCCGAAGCAGAUUGCCGAACAUGCUGUUUGGAGUCGAAAGACUUUUGGCCAAAUAUGAUGUAA_UGCACAUAUAUUUGUAGAC___AGCACAUGAAGUGUGUGUUGUUGGUUAAA ...(((((((((((..((....))..)))).....(((((....)))))))))))).............................(((...(((((((.....)))))))....)))... (-25.15 = -25.27 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:57 2006