| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 678,303 – 678,485 |
| Length | 182 |
| Max. P | 0.874367 |

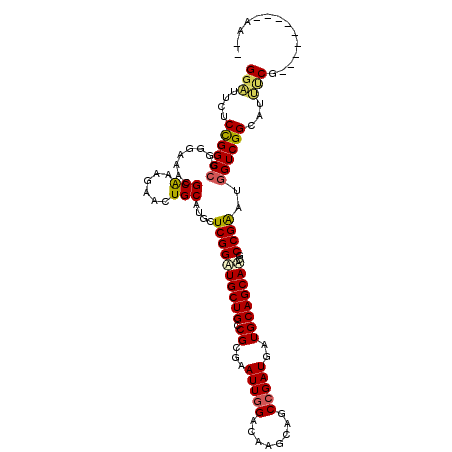
| Location | 678,303 – 678,418 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.93 |
| Mean single sequence MFE | -44.02 |
| Consensus MFE | -30.44 |
| Energy contribution | -30.08 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 678303 115 + 23771897 GGGUUCUCCGGCCGGGGAAACGCAAAGAACUGCAUACUCGGGUGCUGACGCGAAUUGGACAAGCAUCCGAUGAUGCAGCACCGCCGGAUGGUCGGCAUCUCGACCUGCGGAUGAG ..((((((((((.(((....)........((((((..(((((((((....(......)...)))))))))..))))))..)))))))).((((((....))))))...))))... ( -47.80) >DroVir_CAF1 40695 103 + 1 GGAUUGUCCGGCCUGGGAAAGGCGAAGAACUGCAUGCUCGGAUGCUGCCGCGAAUUCGACAAGCAUCCGAUGAUGCAGCAUCGCCGGAUAGUCGGCAUAACGG------------ .(((((((((((((.....))))...((.((((((..(((((((((....((....))...)))))))))..))))))..))..)))))))))..........------------ ( -46.60) >DroPse_CAF1 40694 108 + 1 GGACUCUCCGGCCGUGGAAAGGCGAAGAACUGCAUGCUCGGAUGCUGCCGGGAAUUGGACAUGCAGCCGAUGAUGCAGCAACGCCGAAUGGUCGGCAUUCCG-------GAAACG (((....((((((((.....((((..(..((((((..((((.((((((((.....))).)).))).))))..)))))))..))))..))))))))...)))(-------....). ( -45.20) >DroSec_CAF1 33927 102 + 1 GGGUUCUCCGGCCGGGGAAACGCAAAGAACUGCAUACUCGGGUGCUGACGCGAAUUGGACAGGCAUCCGAUGAUGCAGCACCGCCGGAUGGUCGGCAUCUCG------------- .(..(.((((((.(((....)........((((((..(((((((((....(......)...)))))))))..))))))..)))))))).)..).........------------- ( -40.40) >DroWil_CAF1 13215 106 + 1 GGAUUCUCUGGCCUGGGAAACGCAAAGAACUGCAUGCUCGGAUGCUGUCGCGAAUUGGACAGGCAGCCGAUAAUGCAGCAUCGCCGAAUGGUCGGCAUUUCGGC-------AA-- ...(((((......)))))..((...(((((((((..((((.(((((((........)))).))).))))..))))))....(((((....)))))..))).))-------..-- ( -38.90) >DroPer_CAF1 45120 108 + 1 GGACUUUCCGGCCGUGGAAAGGCGAAGAACUGCAUGCUCGGAUGCUGCCGGGAAUUGGACAUGCAGCCGAUGAUGCAGCAACGCCGAAUGGUCGGCAUUCCG-------GAAACG (((....((((((((.....((((..(..((((((..((((.((((((((.....))).)).))).))))..)))))))..))))..))))))))...)))(-------....). ( -45.20) >consensus GGAUUCUCCGGCCGGGGAAACGCAAAGAACUGCAUGCUCGGAUGCUGCCGCGAAUUGGACAAGCAGCCGAUGAUGCAGCAACGCCGAAUGGUCGGCAUUUCG_________AA__ (((....((((((........(((......)))....((((((((((.((...(((((........)))))..))))))))..))))..))))))...))).............. (-30.44 = -30.08 + -0.36)

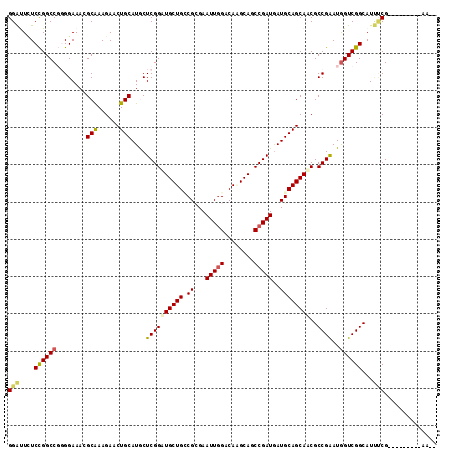
| Location | 678,303 – 678,418 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.93 |
| Mean single sequence MFE | -41.75 |
| Consensus MFE | -32.37 |
| Energy contribution | -32.57 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 678303 115 - 23771897 CUCAUCCGCAGGUCGAGAUGCCGACCAUCCGGCGGUGCUGCAUCAUCGGAUGCUUGUCCAAUUCGCGUCAGCACCCGAGUAUGCAGUUCUUUGCGUUUCCCCGGCCGGAGAACCC .((.......(((((......))))).((((((((((((((......((((....)))).......).))))))).(((.((((((....)))))))))....)))))))).... ( -41.72) >DroVir_CAF1 40695 103 - 1 ------------CCGUUAUGCCGACUAUCCGGCGAUGCUGCAUCAUCGGAUGCUUGUCGAAUUCGCGGCAGCAUCCGAGCAUGCAGUUCUUCGCCUUUCCCAGGCCGGACAAUCC ------------..(((..(((........(((((.(((((((..(((((((((.((((......)))))))))))))..)))))))...))))).......)))..)))..... ( -47.26) >DroPse_CAF1 40694 108 - 1 CGUUUC-------CGGAAUGCCGACCAUUCGGCGUUGCUGCAUCAUCGGCUGCAUGUCCAAUUCCCGGCAGCAUCCGAGCAUGCAGUUCUUCGCCUUUCCACGGCCGGAGAGUCC ..((((-------(((((((((((....))))))))(((((((..((((.(((.((((........))))))).))))..))))))).................))))))).... ( -43.70) >DroSec_CAF1 33927 102 - 1 -------------CGAGAUGCCGACCAUCCGGCGGUGCUGCAUCAUCGGAUGCCUGUCCAAUUCGCGUCAGCACCCGAGUAUGCAGUUCUUUGCGUUUCCCCGGCCGGAGAACCC -------------......((((......))))((((((((((..((((.(((.((.((.....).).))))).))))..)))))))((((((.(((.....)))))))))))). ( -34.50) >DroWil_CAF1 13215 106 - 1 --UU-------GCCGAAAUGCCGACCAUUCGGCGAUGCUGCAUUAUCGGCUGCCUGUCCAAUUCGCGACAGCAUCCGAGCAUGCAGUUCUUUGCGUUUCCCAGGCCAGAGAAUCC --((-------((((((.((.....)))))))))).(((((((..((((.(((.(((((.....).))))))).))))..)))))))((((((.(((.....))))))))).... ( -39.70) >DroPer_CAF1 45120 108 - 1 CGUUUC-------CGGAAUGCCGACCAUUCGGCGUUGCUGCAUCAUCGGCUGCAUGUCCAAUUCCCGGCAGCAUCCGAGCAUGCAGUUCUUCGCCUUUCCACGGCCGGAAAGUCC ..((((-------(((((((((((....))))))))(((((((..((((.(((.((((........))))))).))))..))))))).................))))))).... ( -43.60) >consensus __UU_________CGAAAUGCCGACCAUCCGGCGAUGCUGCAUCAUCGGAUGCAUGUCCAAUUCGCGGCAGCAUCCGAGCAUGCAGUUCUUCGCCUUUCCCCGGCCGGAGAAUCC ..................(((((......)))))..(((((((..((((.(((.((((........))))))).))))..)))))))(((((((.........).)))))).... (-32.37 = -32.57 + 0.20)

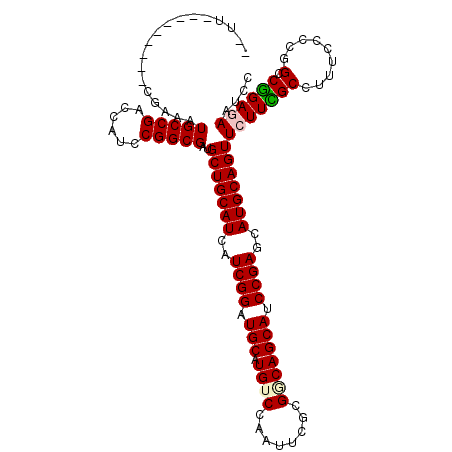
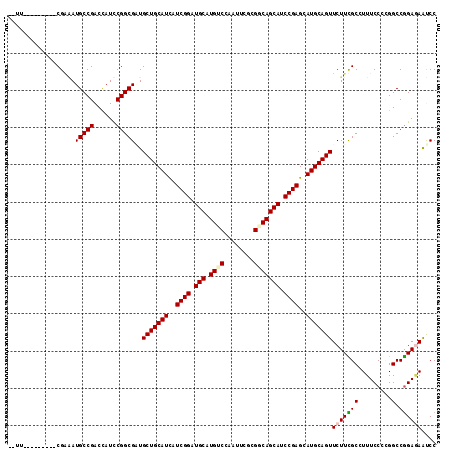
| Location | 678,383 – 678,485 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 72.79 |
| Mean single sequence MFE | -30.89 |
| Consensus MFE | -20.42 |
| Energy contribution | -19.95 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
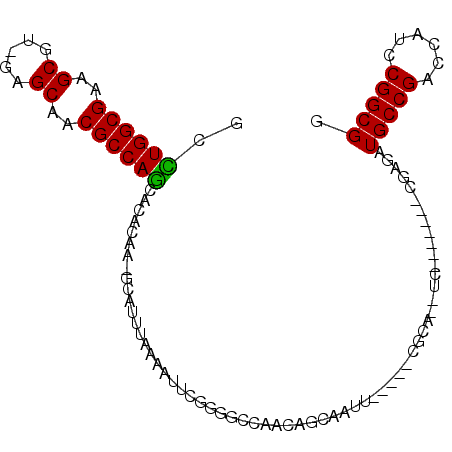
>3L_DroMel_CAF1 678383 102 - 23771897 ACCUGGCGAAGCGU-GAGCAACGCCAGCACUCCA-GCAUUUAAGAUUCGGGGCCAACAGCAAUUUCAUCCUCA--UCCGCAGGUCGAGAUGCCGACCAUCCGGCGG ..((((((..((..-..))..))))))..((((.-.............))))((.......(((((..((((.--...).)))..)))))((((......)))))) ( -32.54) >DroPse_CAF1 40774 92 - 1 GCUUGGCGGCGCAUUUAGCAACGCCAAGACAAAAUUCAUUAAAAAUUCGGGGCAUACAGAACAA-----CGUU--UC-------CGGAAUGCCGACCAUUCGGCGU .(((((((..((.....))..))))))).................(((((((...((.......-----.)))--))-------))))(((((((....))))))) ( -28.40) >DroSec_CAF1 34007 84 - 1 GCCUGGCGAAGCGU-GAGCAACGCCAGCACUCCA-GCAUUUAAGAUUCGGGGCCAACAGCAAUU--------------------CGAGAUGCCGACCAUCCGGCGG ((.(((((..((..-..))..)))))))...((.-..........(((((.((.....))...)--------------------))))..((((......)))))) ( -28.80) >DroYak_CAF1 36067 102 - 1 GCCUGGCGAAGCGU-GAGCAACGCCAGCACACCA-GCAUUUAAGAUUCGGGGCCAACAGCAAUUCCAUCCGCA--UCCGCAGGUCGAGAUGCCGACCAUCCGGCGG ((((((((..((..-..))..)))).((......-))............)))).....((..........)).--.((((.(((((......))))).....)))) ( -33.30) >DroAna_CAF1 35268 96 - 1 GCUUGGCGAAGCGU-CAGCAACGCCAGCUCCUAA-AGAUUUAAAAUUCGGGGCCAUCAGCA--------CGCAAUUCCGCAGCCCGAGAUGCCGACCAUUCGGCGU (((.((((..((..-..))..)))))))......-..........((((((((.....)).--------.((......))..))))))(((((((....))))))) ( -33.90) >DroPer_CAF1 45200 92 - 1 GCUUGGCGGCGCAUUUAGCAACGCCAAGACAAAAUUCAUUAAAAAUUCGGGGCAUACAGAACAA-----CGUU--UC-------CGGAAUGCCGACCAUUCGGCGU .(((((((..((.....))..))))))).................(((((((...((.......-----.)))--))-------))))(((((((....))))))) ( -28.40) >consensus GCCUGGCGAAGCGU_GAGCAACGCCAGCACACAA_GCAUUUAAAAUUCGGGGCCAACAGCAAUU_____CGCA__UC_______CGAGAUGCCGACCAUCCGGCGG ..((((((..((.....))..))))))..............................................................(((((......))))). (-20.42 = -19.95 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:29 2006