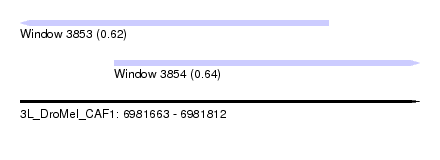
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,981,663 – 6,981,812 |
| Length | 149 |
| Max. P | 0.644166 |

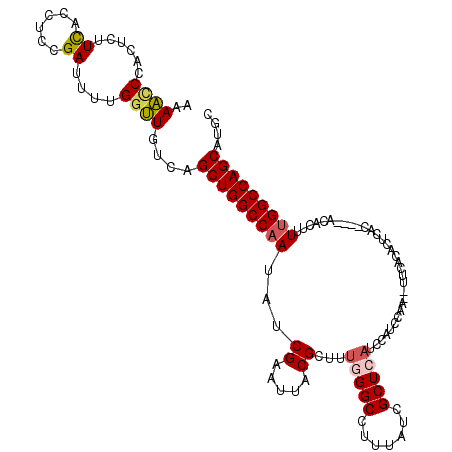
| Location | 6,981,663 – 6,981,778 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.40 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -23.38 |
| Energy contribution | -23.30 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6981663 115 - 23771897 AUGGAUGAGCGAUAAAGGCCCAAAGCGUAAUUCGAUAUUGGCCAGCUGACAACCAAAAUCGGAGGUGAUGAGUGGGUUUUCGUUGCAGAGGCAUAA-----UAAUUAUAAAUUGAUGAUA ........((((..((((((((...(((..((((((.((((...........)))).))))))....)))..))))))))..))))..........-----..(((((......))))). ( -29.10) >DroSec_CAF1 51497 115 - 1 AUGGAAGAGCGAUAAAGGCCCAAAGCGUAAUUCGAUAUUGGCCAGCUGACAACCAAAAUCGGAGGUGAGGAGUGGAUUUUGGUGGCAAACAAAUAA-----UAAUUAUAAAUGUAUGAUA ........((......((((.....((.....)).....))))........(((((((((.(..........).))))))))).))..........-----..((((((....)))))). ( -21.30) >DroSim_CAF1 51273 115 - 1 AUGGAUGAGCGAUAAAGGCCUAAAGCGUAAUUCGAUAUUGGCCAGCUGACAACCAAAAUCGGAGGUAAAGAGUGGGUUUUGGUUGCAGAGACAUAA-----UAAUUAUAAAUGGAUGAUA ....(((.........((((.....((.....)).....))))..(((.(((((((((((.(..........).))))))))))))))...)))..-----..(((((......))))). ( -26.60) >DroEre_CAF1 51306 115 - 1 CUGGAUGAGCGAUAAAGGCCUAAAGCGUAAUUCGAUAUUGGCCAGCUGGCAGUCAAAUUCGGAGGUGAAGAGUGGGCUUUGGUUGCAGAGACAAAA-----UAAUUAUAAAUGUAUGAAA .((..(..(((((.((((((((.....((.(((((..((((((....)))...)))..)))))..)).....)))))))).)))))..)..))...-----................... ( -26.40) >DroYak_CAF1 53141 120 - 1 AUGGAUGAGCGAUAAAGGCCCAAAGCGUAAUUCGAUAUUGGCCAGCUGGCAACCAAAAUCGGAGGUGAAGAGUGGGUUUCGAUUGCAGAGAUAUAAUACCAUAAUUAUAAAUGUAUGAUA ........(((((..(((((((.....((.((((((.((((((....)))...))).))))))..)).....)))))))..))))).....((((((......))))))........... ( -28.60) >consensus AUGGAUGAGCGAUAAAGGCCCAAAGCGUAAUUCGAUAUUGGCCAGCUGACAACCAAAAUCGGAGGUGAAGAGUGGGUUUUGGUUGCAGAGACAUAA_____UAAUUAUAAAUGUAUGAUA ........(((((.((((((((.....((.((((((.((((...........)))).))))))..)).....)))))))).))))).................................. (-23.38 = -23.30 + -0.08)

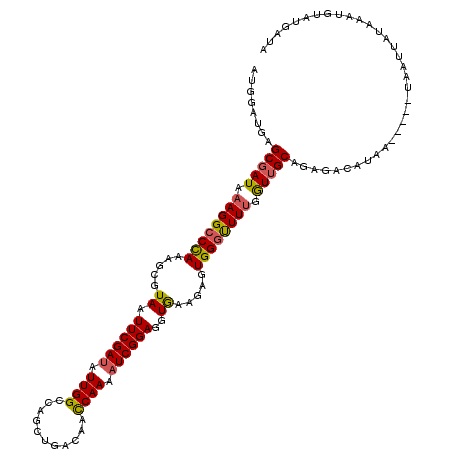
| Location | 6,981,698 – 6,981,812 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.39 |
| Mean single sequence MFE | -21.54 |
| Consensus MFE | -17.62 |
| Energy contribution | -18.02 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6981698 114 + 23771897 AAAACCCACUCAUCACCUCCGAUUUUGGUUGUCAGCUGGCCAAUAUCGAAUUACGCUUUGGGCCUUUAUCGCUCAUCCAUCCAA--UUCACACUCAC----ACACUUUUGGCCAGCAUGC ..((((.....(((......)))...))))....(((((((((....(((((......(((((.......))))).......))--)))........----......))))))))).... ( -24.29) >DroSec_CAF1 51532 118 + 1 AAAAUCCACUCCUCACCUCCGAUUUUGGUUGUCAGCUGGCCAAUAUCGAAUUACGCUUUGGGCCUUUAUCGCUCUUCCAUCCGA--UUCACACUCACACACACACUUUUGGCCAGCAUGC ..............((..(((....)))..))..(((((((((....(((((.......((((.......))))........))--)))..................))))))))).... ( -21.71) >DroSim_CAF1 51308 118 + 1 AAAACCCACUCUUUACCUCCGAUUUUGGUUGUCAGCUGGCCAAUAUCGAAUUACGCUUUAGGCCUUUAUCGCUCAUCCAUCCGA--UUCACACUCACACACACACUUUUGGCCAGCAUGC ..............((..(((....)))..))..(((((((((...........((.....))....((((..........)))--)....................))))))))).... ( -20.00) >DroEre_CAF1 51341 114 + 1 AAAGCCCACUCUUCACCUCCGAAUUUGACUGCCAGCUGGCCAAUAUCGAAUUACGCUUUAGGCCUUUAUCGCUCAUCCAGCCAA--UCAACACUCAC----ACACUUUCGGCCAGCAUGC ..................................(((((((......((.....((.....)).....))(((.....)))...--...........----........))))))).... ( -18.80) >DroYak_CAF1 53181 116 + 1 GAAACCCACUCUUCACCUCCGAUUUUGGUUGCCAGCUGGCCAAUAUCGAAUUACGCUUUGGGCCUUUAUCGCUCAUCCAUCCAAAUCCAACACUCAC----ACACUUUUGGCCAGCAUGC ..((((......((......))....))))....(((((((((...((.....))...(((((.......)))))......................----......))))))))).... ( -22.90) >consensus AAAACCCACUCUUCACCUCCGAUUUUGGUUGUCAGCUGGCCAAUAUCGAAUUACGCUUUGGGCCUUUAUCGCUCAUCCAUCCAA__UUCACACUCAC____ACACUUUUGGCCAGCAUGC ..((((......((......))....))))....(((((((((...((.....))...(((((.......)))))................................))))))))).... (-17.62 = -18.02 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:48 2006