
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,980,811 – 6,980,986 |
| Length | 175 |
| Max. P | 0.999944 |

| Location | 6,980,811 – 6,980,908 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.48 |
| Mean single sequence MFE | -13.74 |
| Consensus MFE | -10.14 |
| Energy contribution | -11.14 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6980811 97 - 23771897 -AAAAACUGAAUCGCCAGACACAAAAAAUGUUCAAAAAA----CAAAAAGUAUAAAUAUGCAGCGAUCUCGGUUUUGCUUUCCAUAA------------CCAA------UUACUUUUACU -.(((((((((((((..((.(((.....)))))......----......((((....)))).))))).))))))))...........------------....------........... ( -15.90) >DroSec_CAF1 50666 98 - 1 UAAAAACUGAAUCGCCAGACACAAAAAAUGUUCAAAAAA----CAAAAAGUAUAAAUAUGCACCGAUCUCGGUUUUGCUUUCCACAA------------CCAU------UUACUUUUACU ..((((((((((((...((.(((.....)))))......----......((((....))))..)))).))))))))...........------------....------........... ( -12.20) >DroSim_CAF1 50418 98 - 1 UAAAAACUGAAUCGCCAGACACAAAAAAUGUUCAAAAAA----CAAAAAGUAUAAAUAUGCAGCGAUCUCGGUUUUGCUUCCCAUAC------------CCAU------UUACUUUUACU ..(((((((((((((..((.(((.....)))))......----......((((....)))).))))).))))))))...........------------....------........... ( -15.90) >DroEre_CAF1 50484 107 - 1 -AAAAACUGAAUCGCCAGGCGCAGAAAACGUUCAUAAAAGAUAUAAAAAGUACAAAUAUGCAGCGAUCUCCGUUUUGUUUCCCACUCCCACUCCCA------A------UUACAUUUACU -.(((((.(((((((..((((.......)))).................(((......))).))))).)).)))))....................------.------........... ( -10.80) >DroYak_CAF1 52319 113 - 1 -AAAAACUGAAUCGCCAGACAGAAAAAACAUUCAUAA------CAAAAAGUAUAAAUAUGCACCGAUCUCGGUUGUGGUACCCACUCCUACUCCUACUCCCAUUUACUUUUACAUUUACU -.....(((......)))...(((......)))....------..(((((((.........((((....)))).((((...))))...................)))))))......... ( -13.90) >consensus _AAAAACUGAAUCGCCAGACACAAAAAAUGUUCAAAAAA____CAAAAAGUAUAAAUAUGCAGCGAUCUCGGUUUUGCUUCCCACAC____________CCAU______UUACUUUUACU ..(((((((((((((..((((.......)))).................((((....)))).))))).))))))))............................................ (-10.14 = -11.14 + 1.00)

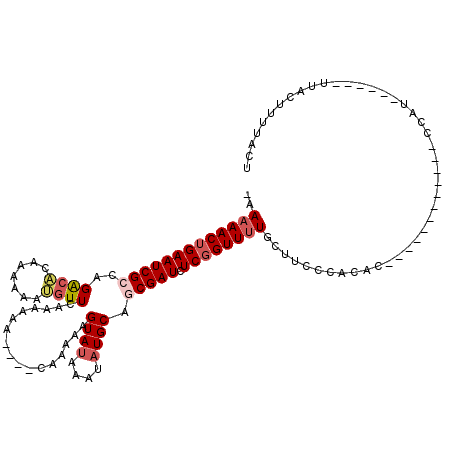
| Location | 6,980,833 – 6,980,947 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.03 |
| Mean single sequence MFE | -21.44 |
| Consensus MFE | -18.02 |
| Energy contribution | -19.02 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6980833 114 - 23771897 UUAAUUCCCUUGAUAAUGAUGCCGGCAAAAUGGCAACAU--AAAAACUGAAUCGCCAGACACAAAAAAUGUUCAAAAAA----CAAAAAGUAUAAAUAUGCAGCGAUCUCGGUUUUGCUU ...............(((.(((((......))))).)))--.(((((((((((((..((.(((.....)))))......----......((((....)))).))))).)))))))).... ( -24.20) >DroSec_CAF1 50688 116 - 1 UUAAUUCACUUGAUAAUGAUGCCGGCAAAAUGGCAACAUAUAAAAACUGAAUCGCCAGACACAAAAAAUGUUCAAAAAA----CAAAAAGUAUAAAUAUGCACCGAUCUCGGUUUUGCUU ...............(((.(((((......))))).)))...((((((((((((...((.(((.....)))))......----......((((....))))..)))).)))))))).... ( -20.50) >DroSim_CAF1 50440 116 - 1 UUAAUUCCCUUGAUAAUGAUGCCGGCAAAAUGGCAACAUAUAAAAACUGAAUCGCCAGACACAAAAAAUGUUCAAAAAA----CAAAAAGUAUAAAUAUGCAGCGAUCUCGGUUUUGCUU ...............(((.(((((......))))).)))...(((((((((((((..((.(((.....)))))......----......((((....)))).))))).)))))))).... ( -24.20) >DroEre_CAF1 50512 118 - 1 UUAAUUCCCUUGAUAAUGAUGCCGGCAAAAUGGCAACAU--AAAAACUGAAUCGCCAGGCGCAGAAAACGUUCAUAAAAGAUAUAAAAAGUACAAAUAUGCAGCGAUCUCCGUUUUGUUU ...............(((.(((((......))))).)))--.(((((.(((((((..((((.......)))).................(((......))).))))).)).))))).... ( -19.10) >DroYak_CAF1 52359 112 - 1 UUAAUUCCCUUGAUAAUGAUGCCGGCAAAAUGGCAACAU--AAAAACUGAAUCGCCAGACAGAAAAAACAUUCAUAA------CAAAAAGUAUAAAUAUGCACCGAUCUCGGUUGUGGUA .......((......(((.(((((......))))).)))--...((((((((((.......(((......)))....------......((((....))))..)))).))))))..)).. ( -19.20) >consensus UUAAUUCCCUUGAUAAUGAUGCCGGCAAAAUGGCAACAU__AAAAACUGAAUCGCCAGACACAAAAAAUGUUCAAAAAA____CAAAAAGUAUAAAUAUGCAGCGAUCUCGGUUUUGCUU ...............(((.(((((......))))).)))...(((((((((((((..((((.......)))).................((((....)))).))))).)))))))).... (-18.02 = -19.02 + 1.00)

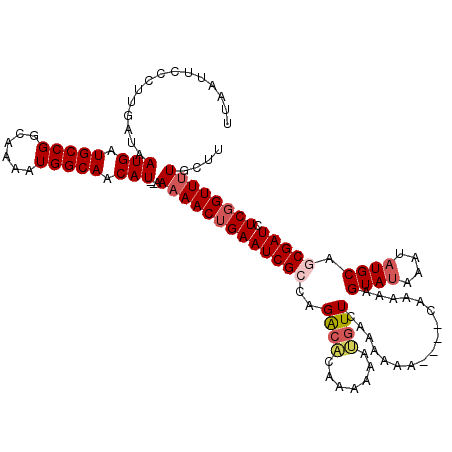
| Location | 6,980,870 – 6,980,986 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.46 |
| Mean single sequence MFE | -27.96 |
| Consensus MFE | -24.78 |
| Energy contribution | -25.38 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6980870 116 + 23771897 -UUUUUUGAACAUUUUUUGUGUCUGGCGAUUCAGUUUUU--AUGUUGCCAUUUUGCCGGCAUCAUUAUCAAGGGAAUUAACCCAAAUGCCAAACACAGAGAUGAUGUUUUUUUU-UGUGC -......(((((((((((((((.(((((...........--(((.((((........)))).)))......(((......)))...))))).))))))))..))))))).....-..... ( -31.40) >DroSec_CAF1 50725 114 + 1 -UUUUUUGAACAUUUUUUGUGUCUGGCGAUUCAGUUUUUAUAUGUUGCCAUUUUGCCGGCAUCAUUAUCAAGUGAAUUAACCCAAAUGCCAAACACAGAGAUGAUGUUUUUUUGG----- -......(((((((((((((((.((((((((((.((..((.(((.((((........)))).)))))..)).)))))).........)))).))))))))..)))))))......----- ( -30.50) >DroSim_CAF1 50477 119 + 1 -UUUUUUGAACAUUUUUUGUGUCUGGCGAUUCAGUUUUUAUAUGUUGCCAUUUUGCCGGCAUCAUUAUCAAGGGAAUUAACCCAAAUGCCAAACACAGAGAUGAUGUUUUUUUUGUGUGU -......(((((((((((((((.(((((.............(((.((((........)))).)))......(((......)))...))))).))))))))..)))))))........... ( -31.40) >DroEre_CAF1 50552 113 + 1 CUUUUAUGAACGUUUUCUGCGCCUGGCGAUUCAGUUUUU--AUGUUGCCAUUUUGCCGGCAUCAUUAUCAAGGGAAUUAACCCAAAUGCUAAACACAGAGACGCUGAUUUUUU-----GU ..........(((((.(((.(..(((((...........--(((.((((........)))).)))......(((......)))...)))))..).))))))))..........-----.. ( -23.70) >DroYak_CAF1 52396 111 + 1 ---UUAUGAAUGUUUUUUCUGUCUGGCGAUUCAGUUUUU--AUGUUGCCAUUUUGCCGGCAUCAUUAUCAAGGGAAUUAACCCAAAUGCUAAACACAGAGAUGAUGCUUUUUUG----GU ---........(((.(((((((.(((((...........--(((.((((........)))).)))......(((......)))...)))))...))))))).))).........----.. ( -22.80) >consensus _UUUUUUGAACAUUUUUUGUGUCUGGCGAUUCAGUUUUU__AUGUUGCCAUUUUGCCGGCAUCAUUAUCAAGGGAAUUAACCCAAAUGCCAAACACAGAGAUGAUGUUUUUUUG____GU .......(((((((((((((((.(((((.............(((.((((........)))).)))......(((......)))...))))).)))))))))..))))))........... (-24.78 = -25.38 + 0.60)



| Location | 6,980,870 – 6,980,986 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.46 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -25.00 |
| Energy contribution | -25.28 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.73 |
| SVM RNA-class probability | 0.999944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6980870 116 - 23771897 GCACA-AAAAAAAACAUCAUCUCUGUGUUUGGCAUUUGGGUUAAUUCCCUUGAUAAUGAUGCCGGCAAAAUGGCAACAU--AAAAACUGAAUCGCCAGACACAAAAAAUGUUCAAAAAA- .....-......(((((......(((((((((((((((((......)))......(((.(((((......))))).)))--.......)))).))))))))))....))))).......- ( -30.20) >DroSec_CAF1 50725 114 - 1 -----CCAAAAAAACAUCAUCUCUGUGUUUGGCAUUUGGGUUAAUUCACUUGAUAAUGAUGCCGGCAAAAUGGCAACAUAUAAAAACUGAAUCGCCAGACACAAAAAAUGUUCAAAAAA- -----.......(((((......(((((((((((((..((((((.....))))).(((.(((((......))))).))).......)..))).))))))))))....))))).......- ( -29.80) >DroSim_CAF1 50477 119 - 1 ACACACAAAAAAAACAUCAUCUCUGUGUUUGGCAUUUGGGUUAAUUCCCUUGAUAAUGAUGCCGGCAAAAUGGCAACAUAUAAAAACUGAAUCGCCAGACACAAAAAAUGUUCAAAAAA- ............(((((......(((((((((((((((((......)))......(((.(((((......))))).))).........)))).))))))))))....))))).......- ( -30.20) >DroEre_CAF1 50552 113 - 1 AC-----AAAAAAUCAGCGUCUCUGUGUUUAGCAUUUGGGUUAAUUCCCUUGAUAAUGAUGCCGGCAAAAUGGCAACAU--AAAAACUGAAUCGCCAGGCGCAGAAAACGUUCAUAAAAG ..-----........(((((.(((((((((.(((((((((......)))......(((.(((((......))))).)))--.......)))).)).)))))))))..)))))........ ( -29.80) >DroYak_CAF1 52396 111 - 1 AC----CAAAAAAGCAUCAUCUCUGUGUUUAGCAUUUGGGUUAAUUCCCUUGAUAAUGAUGCCGGCAAAAUGGCAACAU--AAAAACUGAAUCGCCAGACAGAAAAAACAUUCAUAA--- ..----...............(((((((((((((...(((......))).))...(((.(((((......))))).)))--.....))))).)).))))..(((......)))....--- ( -19.30) >consensus AC____AAAAAAAACAUCAUCUCUGUGUUUGGCAUUUGGGUUAAUUCCCUUGAUAAUGAUGCCGGCAAAAUGGCAACAU__AAAAACUGAAUCGCCAGACACAAAAAAUGUUCAAAAAA_ .......................(((((((((((((..((((((.....))))).(((.(((((......))))).))).......)..))).))))))))))................. (-25.00 = -25.28 + 0.28)

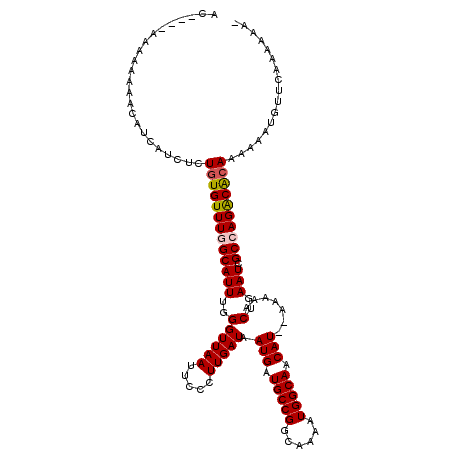
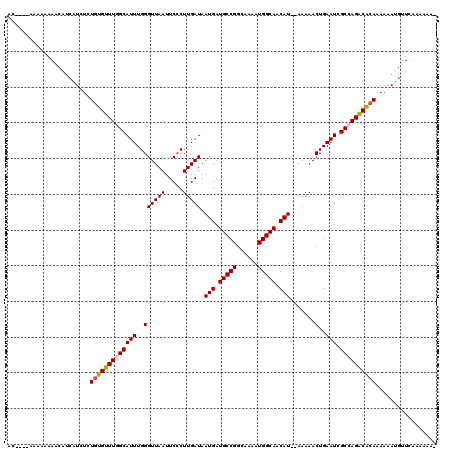
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:45 2006