
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,980,196 – 6,980,463 |
| Length | 267 |
| Max. P | 0.998920 |

| Location | 6,980,196 – 6,980,314 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.86 |
| Mean single sequence MFE | -31.21 |
| Consensus MFE | -22.96 |
| Energy contribution | -24.40 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6980196 118 + 23771897 AAGUUCGAUGGGACACGCAUGGCCACAGCGGCAGAUCUGGAUAUCCCAAU-UCCAUUCGAAUUCCCAUUCUGAAUCUGUAUCCGAGAC-GCCUCAUGAUGCACAUUUCAUUGAACUUUUU (((((((((((((..((.((((((.....))).((..(((.....)))..-))))).))...))))))).((((..((((((.(((..-..)))..))))))...))))..))))))... ( -34.50) >DroSec_CAF1 50060 118 + 1 AAGUUCGAUGGGACACGCACGGCCACAGCGGCAGAUCUGGAUAUCCCAAU-CCCAAUCCCAUUCCCAUUCUGAAUCUGUAUCCGAGAC-GCCUCAUGAUGCACAUUUCAUUGAACUUUUU (((((((((((((........(((.....))).(((..((((......))-))..)))....))))))).((((..((((((.(((..-..)))..))))))...))))..))))))... ( -33.20) >DroSim_CAF1 49802 118 + 1 AAGUUCGAUGGGACACGCACGGCCACAGCGGCAGAUCUGGAUAUCCCAAA-CCCAUUCCAAUUCCCAUUCUGAAUCUGUAUCCGAGAC-GCCUCAUGAUGCACAUUUCAUUGAACUUUUU ((((((((((..(...(((.((..((((...((((...(((.........-...........)))...))))...))))..))(((..-..)))....)))...)..))))))))))... ( -31.45) >DroEre_CAF1 49996 107 + 1 AAGUUCGAUGCGACACGCACGGCCACAGCCUCAGAUCUGGAU-------------AUCCCAGUCCCAUUCCGAUUCCCAUUUCGAAUCUGCCUCAUGAUGCACAUUUCAUUGAACUUUUU ((((((((((.((...(((.(((....))).(((((((((..-------------...))))........(((........))).)))))........)))...)).))))))))))... ( -25.60) >DroYak_CAF1 51667 119 + 1 AAGUUCGAUGGGACACGCACGGCCACAGCCUCAGAUCUGGAUCUGUGGAUAUUCAAUCCCAAUCCCAUUACGAAUCUGUAUCCGAAAC-GCCUCAUGAUGCACAUUUCAUUGAACUUUUU (((((((((((((.......(((....))).(((((....))))).((((.....))))...)))))))..(((..((((((.((...-...))..))))))...)))...))))))... ( -31.30) >consensus AAGUUCGAUGGGACACGCACGGCCACAGCGGCAGAUCUGGAUAUCCCAAU_CCCAAUCCCAUUCCCAUUCUGAAUCUGUAUCCGAGAC_GCCUCAUGAUGCACAUUUCAUUGAACUUUUU ((((((((((..(...(((.((..((((...((((...(((.....................)))...))))...))))..))(((.....)))....)))...)..))))))))))... (-22.96 = -24.40 + 1.44)

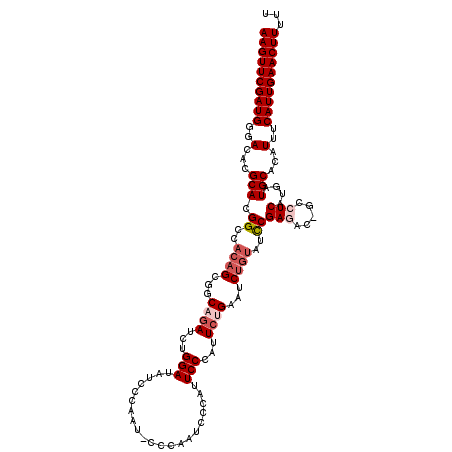

| Location | 6,980,196 – 6,980,314 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.86 |
| Mean single sequence MFE | -35.72 |
| Consensus MFE | -27.88 |
| Energy contribution | -28.48 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6980196 118 - 23771897 AAAAAGUUCAAUGAAAUGUGCAUCAUGAGGC-GUCUCGGAUACAGAUUCAGAAUGGGAAUUCGAAUGGA-AUUGGGAUAUCCAGAUCUGCCGCUGUGGCCAUGCGUGUCCCAUCGAACUU ...((((((.(((..(..(((((...(((..-..)))((.(((((...((((.((((.((((.(((...-))).)))).))))..))))...))))).)))))))..)..))).)))))) ( -36.90) >DroSec_CAF1 50060 118 - 1 AAAAAGUUCAAUGAAAUGUGCAUCAUGAGGC-GUCUCGGAUACAGAUUCAGAAUGGGAAUGGGAUUGGG-AUUGGGAUAUCCAGAUCUGCCGCUGUGGCCGUGCGUGUCCCAUCGAACUU ...((((((..((((.(.((.(((.((((..-..))))))).)).)))))((.(((((((((((((.((-((......)))).)))))(((.....)))....))).))))))))))))) ( -34.80) >DroSim_CAF1 49802 118 - 1 AAAAAGUUCAAUGAAAUGUGCAUCAUGAGGC-GUCUCGGAUACAGAUUCAGAAUGGGAAUUGGAAUGGG-UUUGGGAUAUCCAGAUCUGCCGCUGUGGCCGUGCGUGUCCCAUCGAACUU ...((((((.(((..(..(((((...(((..-..)))((.(((((((((.......)))).((...(((-(((((.....)))))))).)).))))).)))))))..)..))).)))))) ( -35.30) >DroEre_CAF1 49996 107 - 1 AAAAAGUUCAAUGAAAUGUGCAUCAUGAGGCAGAUUCGAAAUGGGAAUCGGAAUGGGACUGGGAU-------------AUCCAGAUCUGAGGCUGUGGCCGUGCGUGUCGCAUCGAACUU ...((((((.(((..(..(((((.....(((.(((((.......)))))((..(.(((((((...-------------..)))).))).)..))...))))))))..)..))).)))))) ( -33.70) >DroYak_CAF1 51667 119 - 1 AAAAAGUUCAAUGAAAUGUGCAUCAUGAGGC-GUUUCGGAUACAGAUUCGUAAUGGGAUUGGGAUUGAAUAUCCACAGAUCCAGAUCUGAGGCUGUGGCCGUGCGUGUCCCAUCGAACUU ...((((((.(((..(((.....)))((.((-((..(((.(((((.((((.....(((((.((((.....))))...))))).....)))).))))).))).)))).)).))).)))))) ( -37.90) >consensus AAAAAGUUCAAUGAAAUGUGCAUCAUGAGGC_GUCUCGGAUACAGAUUCAGAAUGGGAAUGGGAUUGGA_AUUGGGAUAUCCAGAUCUGCCGCUGUGGCCGUGCGUGUCCCAUCGAACUU ...((((((.(((..(..(((((...(((.....)))((.(((((...((((.((((.(((..............))).))))..))))...))))).)))))))..)..))).)))))) (-27.88 = -28.48 + 0.60)

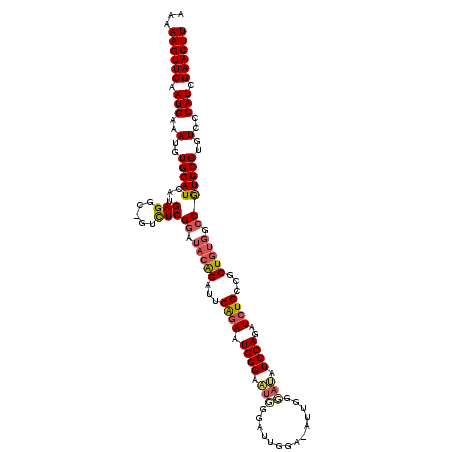
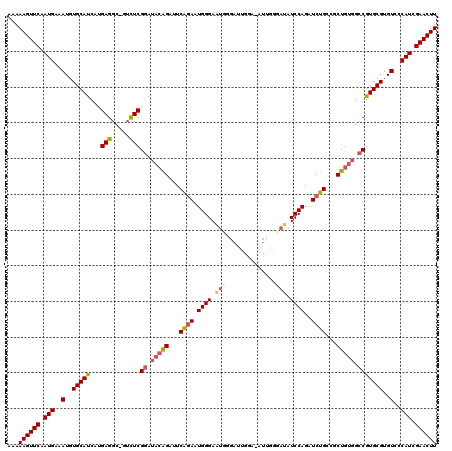
| Location | 6,980,275 – 6,980,387 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.03 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -22.88 |
| Energy contribution | -22.84 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6980275 112 - 23771897 UAGCUGGAAGGCGGCCAUGUGCAACAUGUUGUCGCAAUACCAAAAAU-------AAAAAAAUACGAAGGCGCAGACAGCGAAAAAGUUCAAUGAAAUGUGCAUCAUGAGGC-GUCUCGGA ...(((..(((((.(((((((((.(((((((((((....((......-------.............)).)).))))))(((....)))......))))))).)))).).)-))))))). ( -28.21) >DroSec_CAF1 50139 107 - 1 UAACUGGAAGGCGGCCAUGUGCAACAUGUUGUCGCAAUCCCAAAA------------AAACGACGAAGGCGCAGACAGCGAAAAAGUUCAAUGAAAUGUGCAUCAUGAGGC-GUCUCGGA ...(((..(((((.(((((((((.(((((((((((...((.....------------..........)).)).))))))(((....)))......))))))).)))).).)-))))))). ( -28.86) >DroSim_CAF1 49881 108 - 1 UAGCUGGAAGGCGGCCAUGUGCAACAUGUUGUCGCAAUCCCAAAA-----------AAAAAGUCGAAGGCGCAGACAGCGAAAAAGUUCAAUGAAAUGUGCAUCAUGAGGC-GUCUCGGA ...(((..(((((.(((((((((.(((((((((((...((.....-----------...........)).)).))))))(((....)))......))))))).)))).).)-))))))). ( -28.49) >DroEre_CAF1 50063 110 - 1 UAGCUGGAAGGCGGCCAUGUGCAACAUGUUGUCGCAAUCCAAAA----------AAAUUAAGACGAAGGCGCAGACAGCGAAAAAGUUCAAUGAAAUGUGCAUCAUGAGGCAGAUUCGAA ..((((((..(((((((((.....))))..)))))..)))....----------..........((..(((((..((..(((....)))..))...))))).))....)))......... ( -26.80) >DroYak_CAF1 51747 119 - 1 UAGCUGGAAGGCGGCCAUGUGCAACAUGUUGUCGCAAUCCCAAAAACAAAACAAAAAAAAAAACGAAGGCGCAGACAGCGAAAAAGUUCAAUGAAAUGUGCAUCAUGAGGC-GUUUCGGA ...((((((.((...((((((((.(((((((((((...((...........................)).)).))))))(((....)))......))))))).))))..))-.)))))). ( -26.93) >consensus UAGCUGGAAGGCGGCCAUGUGCAACAUGUUGUCGCAAUCCCAAAA_________AAAAAAAGACGAAGGCGCAGACAGCGAAAAAGUUCAAUGAAAUGUGCAUCAUGAGGC_GUCUCGGA .....(((..(((((((((.....))))..)))))..)))........................((..(((((..((..(((....)))..))...))))).)).((((.....)))).. (-22.88 = -22.84 + -0.04)

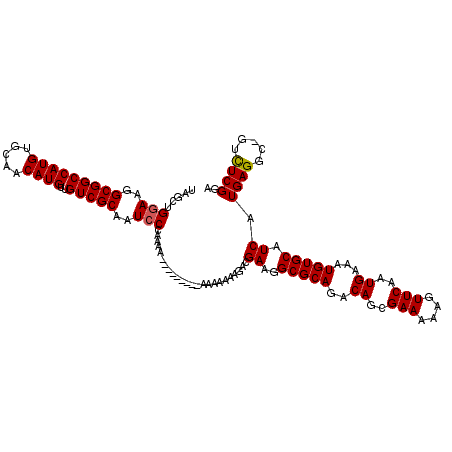
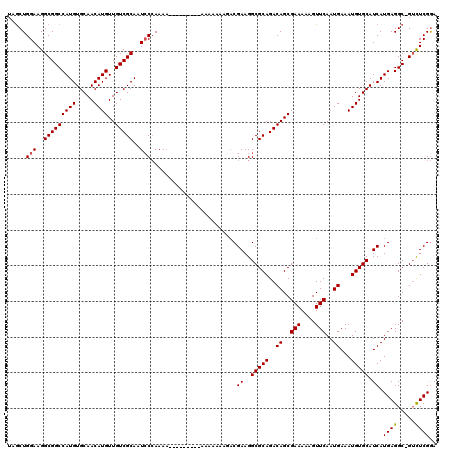
| Location | 6,980,314 – 6,980,427 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.61 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -28.36 |
| Energy contribution | -28.04 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6980314 113 + 23771897 CGCUGUCUGCGCCUUCGUAUUUUUUU-------AUUUUUGGUAUUGCGACAACAUGUUGCACAUGGCCGCCUUCCAGCUAAGUGCAAUAUGAGUAGCACUCGUCGCUUAGAGAUCAUUUA .(.(.((((.(((...(((......)-------))....)))...(((((...(((((((((.((((.(.....).)))).)))))))))(((.....)))))))).)))).).)..... ( -30.70) >DroSec_CAF1 50178 108 + 1 CGCUGUCUGCGCCUUCGUCGUUU------------UUUUGGGAUUGCGACAACAUGUUGCACAUGGCCGCCUUCCAGUUAAGUGCAAUAUGAGUAGCACUCGUCGCUUAGAGAUUAUUUA .((((.((........(((((..------------((....))..)))))..((((((((((.((((.(.....).)))).)))))))))))))))).(((........)))........ ( -30.50) >DroSim_CAF1 49920 109 + 1 CGCUGUCUGCGCCUUCGACUUUUU-----------UUUUGGGAUUGCGACAACAUGUUGCACAUGGCCGCCUUCCAGCUAAGUGCAAUAUGAGUAGCACUCGUCGCUUAGAGAUUAUUUA .((((((.(((..(((((......-----------..)))))..))))))..((((((((((.((((.(.....).)))).))))))))))...))).(((........)))........ ( -30.70) >DroEre_CAF1 50103 110 + 1 CGCUGUCUGCGCCUUCGUCUUAAUUU----------UUUUGGAUUGCGACAACAUGUUGCACAUGGCCGCCUUCCAGCUAAGUGCAAUAUGAGCAGCACUCGUCGCUUAGAGAUUAUUUA .(((((..........(((.((((((----------....)))))).)))..((((((((((.((((.(.....).)))).)))))))))).))))).(((........)))........ ( -33.60) >DroYak_CAF1 51786 120 + 1 CGCUGUCUGCGCCUUCGUUUUUUUUUUUGUUUUGUUUUUGGGAUUGCGACAACAUGUUGCACAUGGCCGCCUUCCAGCUAAGUGCAAUGUGAGUAGCACUCGUCGCUUAGAGAUUAUUUA (((.....)))......................((((((((....(((((...(((((((((.((((.(.....).)))).)))))))))(((.....))))))))))))))))...... ( -29.10) >consensus CGCUGUCUGCGCCUUCGUCUUUUUUU_________UUUUGGGAUUGCGACAACAUGUUGCACAUGGCCGCCUUCCAGCUAAGUGCAAUAUGAGUAGCACUCGUCGCUUAGAGAUUAUUUA (((.....)))..................................(((((...(((((((((.((((.(.....).)))).)))))))))(((.....)))))))).............. (-28.36 = -28.04 + -0.32)


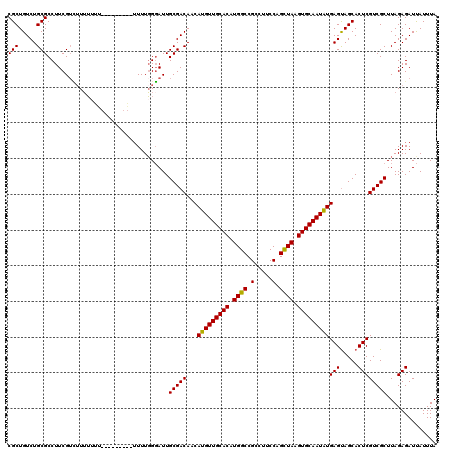
| Location | 6,980,347 – 6,980,463 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.80 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -31.40 |
| Energy contribution | -31.32 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6980347 116 + 23771897 GUAUUGCGACAACAUGUUGCACAUGGCCGCCUUCCAGCUAAGUGCAAUAUGAGUAGCACUCGUCGCUUAGAGAUCAUUUAAAUCACUAGCCCGCAGCUGUUGCACCAAAA----AAAAAA ....((((((..((((((((((.((((.(.....).)))).))))))))))...(((...((..(((.((.(((.......))).))))).))..)))))))))......----...... ( -32.30) >DroSec_CAF1 50206 120 + 1 GGAUUGCGACAACAUGUUGCACAUGGCCGCCUUCCAGUUAAGUGCAAUAUGAGUAGCACUCGUCGCUUAGAGAUUAUUUAAAUCACUCGCCGGCAGCUGUUGUACAAAAAAAAAAAAAAA (((..(((....((((.....))))..)))..)))......((((((((.(((.....)))((((...((.((((.....)))).))...))))...))))))))............... ( -29.70) >DroSim_CAF1 49949 118 + 1 GGAUUGCGACAACAUGUUGCACAUGGCCGCCUUCCAGCUAAGUGCAAUAUGAGUAGCACUCGUCGCUUAGAGAUUAUUUAAAUCACUCGCCGGCAGCUGUUGCACAAAAAA--AAAAAAA ....(((((((.((((((((((.((((.(.....).)))).))))))))))..........((((...((.((((.....)))).))...))))...))))))).......--....... ( -33.20) >DroEre_CAF1 50133 113 + 1 GGAUUGCGACAACAUGUUGCACAUGGCCGCCUUCCAGCUAAGUGCAAUAUGAGCAGCACUCGUCGCUUAGAGAUUAUUUAAAUCACUUGCCGGCAGCUGUUGCACAAAAAA--CG----- ............((((((((((.((((.(.....).)))).)))))))))).((((((((.((((...((.((((.....)))).))...)))))).))))))........--..----- ( -34.80) >DroYak_CAF1 51826 113 + 1 GGAUUGCGACAACAUGUUGCACAUGGCCGCCUUCCAGCUAAGUGCAAUGUGAGUAGCACUCGUCGCUUAGAGAUUAUUUAAAUCACUUGCCGGCAGCUGUUGCACAAAAAA--UG----- ....(((((((.((((((((((.((((.(.....).)))).))))))))))..........((((...((.((((.....)))).))...))))...))))))).......--..----- ( -32.90) >consensus GGAUUGCGACAACAUGUUGCACAUGGCCGCCUUCCAGCUAAGUGCAAUAUGAGUAGCACUCGUCGCUUAGAGAUUAUUUAAAUCACUCGCCGGCAGCUGUUGCACAAAAAA__AAAAAAA ....(((((((.((((((((((.((((.(.....).)))).))))))))))..........((((...((.((((.....)))).))...))))...)))))))................ (-31.40 = -31.32 + -0.08)


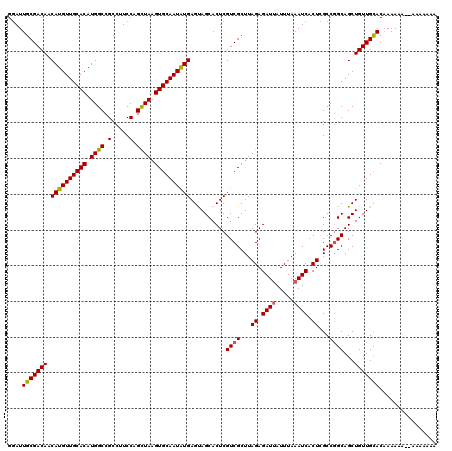
| Location | 6,980,347 – 6,980,463 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.80 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -29.70 |
| Energy contribution | -30.34 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6980347 116 - 23771897 UUUUUU----UUUUGGUGCAACAGCUGCGGGCUAGUGAUUUAAAUGAUCUCUAAGCGACGAGUGCUACUCAUAUUGCACUUAGCUGGAAGGCGGCCAUGUGCAACAUGUUGUCGCAAUAC ......----...((((((..((((((((.(((((.((((.....)))).)).)))..))(((((..........)))))))))))....)).))))(((((((....))).)))).... ( -33.40) >DroSec_CAF1 50206 120 - 1 UUUUUUUUUUUUUUUGUACAACAGCUGCCGGCGAGUGAUUUAAAUAAUCUCUAAGCGACGAGUGCUACUCAUAUUGCACUUAACUGGAAGGCGGCCAUGUGCAACAUGUUGUCGCAAUCC .............(((((((((.((((((..(.(((((((.....))))..........((((((..........)))))).))).)..)))))).(((.....)))))))).))))... ( -31.70) >DroSim_CAF1 49949 118 - 1 UUUUUUU--UUUUUUGUGCAACAGCUGCCGGCGAGUGAUUUAAAUAAUCUCUAAGCGACGAGUGCUACUCAUAUUGCACUUAGCUGGAAGGCGGCCAUGUGCAACAUGUUGUCGCAAUCC .......--....(((((((((.((((((..(.((.((((.....)))).)).(((...((((((..........)))))).))).)..)))))).(((.....))))))).)))))... ( -34.20) >DroEre_CAF1 50133 113 - 1 -----CG--UUUUUUGUGCAACAGCUGCCGGCAAGUGAUUUAAAUAAUCUCUAAGCGACGAGUGCUGCUCAUAUUGCACUUAGCUGGAAGGCGGCCAUGUGCAACAUGUUGUCGCAAUCC -----..--....(((((((((.((((((..(.((.((((.....)))).)).(((...((((((..........)))))).))).)..)))))).(((.....))))))).)))))... ( -34.10) >DroYak_CAF1 51826 113 - 1 -----CA--UUUUUUGUGCAACAGCUGCCGGCAAGUGAUUUAAAUAAUCUCUAAGCGACGAGUGCUACUCACAUUGCACUUAGCUGGAAGGCGGCCAUGUGCAACAUGUUGUCGCAAUCC -----..--....(((((((((.((((((..(.((.((((.....)))).)).(((...((((((..........)))))).))).)..)))))).(((.....))))))).)))))... ( -34.10) >consensus UUUUUUU__UUUUUUGUGCAACAGCUGCCGGCAAGUGAUUUAAAUAAUCUCUAAGCGACGAGUGCUACUCAUAUUGCACUUAGCUGGAAGGCGGCCAUGUGCAACAUGUUGUCGCAAUCC .............(((((((((.((((((....((.((((.....)))).)).(((...((((((..........)))))).)))....)))))).(((.....))))))).)))))... (-29.70 = -30.34 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:41 2006