
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,979,453 – 6,979,613 |
| Length | 160 |
| Max. P | 0.999034 |

| Location | 6,979,453 – 6,979,573 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -37.32 |
| Consensus MFE | -29.44 |
| Energy contribution | -28.80 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6979453 120 - 23771897 CAGACGACAGCUGGCGGAAAGACAAUUAUUUGUCCGUCAGUCUGUCCAAUUUUGAUGGAAUCAUUUGUCUGCGAUGACCUUGUGGCAGCAGAUGUAGGACCAACUGCAUGCUUUCCCAUA ((((((((((((((((((.(((......))).)))))))).))))).......((((....)))).))))).........(((((.((((..(((((......)))))))))...))))) ( -39.50) >DroSec_CAF1 49329 120 - 1 CAGACGACAGCUGCCGGGAAGACAAUUAUUUGUCCGUCAGUCUGUCCACUUUUGAUGGAAUCAUUUGUCUGCGAUGACCUUGUGCCAGCAGAUGUAGGGCCAACUAUAUGCUUUCCCAUA (((.(....))))..((((((((((....)))))....(((..((((((.((((.(((...((...(((......)))..))..))).)))).)).))))..))).......)))))... ( -31.40) >DroSim_CAF1 49070 120 - 1 CAGACGACAGCUGCCGGGAAGACAAUUAUUUGUCCGUCAGUCUGUCCACUUUUGAUGGAAUCAUUUGUCUGCGAUGACCUUGUGCCAGCAGAUGUAGGGCCAACUGCAUGCCAUCCCAUA (((.(....))))..((((.(((((....)))))(((((((..((((((.((((.(((...((...(((......)))..))..))).)))).)).))))..)))).)))...))))... ( -34.30) >DroEre_CAF1 49278 120 - 1 CAGACGACAGCUGUCGGGAAGACAAUUACUUGUCCGUCAGUCUGUCCCAUUUUGAUGGAAUCAUUUGUCUGCGAUGACCUUGUGCCCACAGAUGUAGGGCCAACUGCAUGGUCUCCCAUA (((((((((((((.(((((((.......))).)))).))).)))))((((....))))........))))).((.((((.(((((((((....)).)))).....))).))))))..... ( -41.80) >DroYak_CAF1 50926 120 - 1 CGAACGACAGCUGUCGGGAAGACAAUUAUUUGUCCGUCAGUCUGUCCACUUUUGAUGGAAUCAUUUGUCUGCGAUGACCUUGUGCCCACAGAUGUAGGGCCAACUGCAUGGUCUCCCAUA .....(((....)))((((((((((.......((((((((...(....)..)))))))).....)))))).....((((.(((((((((....)).)))).....))).))))))))... ( -39.60) >consensus CAGACGACAGCUGCCGGGAAGACAAUUAUUUGUCCGUCAGUCUGUCCACUUUUGAUGGAAUCAUUUGUCUGCGAUGACCUUGUGCCAGCAGAUGUAGGGCCAACUGCAUGCUCUCCCAUA ((((.((((((((.(((....((((....))))))).))).)))))....))))((((...(((..(((......)))...))).......((((((......))))))......)))). (-29.44 = -28.80 + -0.64)



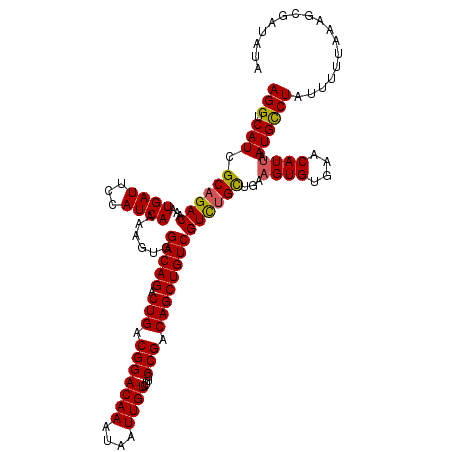
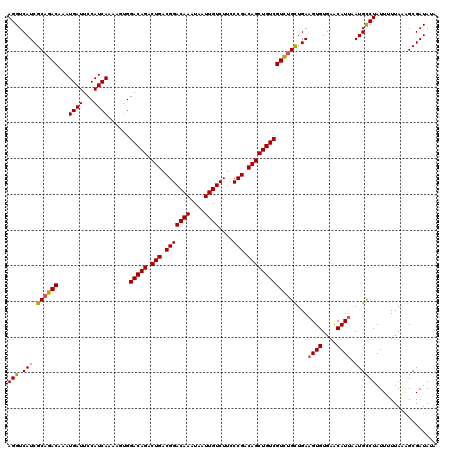
| Location | 6,979,493 – 6,979,613 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.16 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -28.56 |
| Energy contribution | -29.98 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6979493 120 + 23771897 AGGUCAUCGCAGACAAAUGAUUCCAUCAAAAUUGGACAGACUGACGGACAAAUAAUUGUCUUUCCGCCAGCUGUCGUCUGUUGAAGUGUGAACAUUAAUGCCUAUUUUUAAAGCGAUAAA ((((..((((((((...((((...))))......(((((.(((.(((((((....)))....)))).))))))))))))).)))((((....))))...))))................. ( -30.60) >DroSec_CAF1 49369 120 + 1 AGGUCAUCGCAGACAAAUGAUUCCAUCAAAAGUGGACAGACUGACGGACAAAUAAUUGUCUUCCCGGCAGCUGUCGUCUGCUGAAGUGUGAACAUUAAUGCCUAUUUUUAAAGCGAUAUA (((.(((.((((((...((((...))))......(((((.(((.(((((((....))))....))).))))))))))))))...((((....)))).))))))................. ( -32.60) >DroSim_CAF1 49110 120 + 1 AGGUCAUCGCAGACAAAUGAUUCCAUCAAAAGUGGACAGACUGACGGACAAAUAAUUGUCUUCCCGGCAGCUGUCGUCUGCUGAAGUGUGAACAUUAAUGCCUAUUUUUAAAGCGAUAUA (((.(((.((((((...((((...))))......(((((.(((.(((((((....))))....))).))))))))))))))...((((....)))).))))))................. ( -32.60) >DroEre_CAF1 49318 119 + 1 AGGUCAUCGCAGACAAAUGAUUCCAUCAAAAUGGGACAGACUGACGGACAAGUAAUUGUCUUCCCGACAGCUGUCGUCUGCUGAAGUGUGAGCAUU-AUGCCUAUUUUUAAGGCGAUAUA ..(((.((((((((........((((....))))(((((.(((.(((((((....))))....))).)))))))))))))).))((((....))))-..((((.......)))))))... ( -38.70) >DroYak_CAF1 50966 119 + 1 AGGUCAUCGCAGACAAAUGAUUCCAUCAAAAGUGGACAGACUGACGGACAAAUAAUUGUCUUCCCGACAGCUGUCGUUCGCUGAAGUGUGAACAUA-AUGUCUAUUUUUAGAGCGAUAAA .....(((((.......((....))(((((((((((((....(((((........(((((.....))))))))))((((((......))))))...-.))))))))))).)))))))... ( -33.00) >consensus AGGUCAUCGCAGACAAAUGAUUCCAUCAAAAGUGGACAGACUGACGGACAAAUAAUUGUCUUCCCGACAGCUGUCGUCUGCUGAAGUGUGAACAUUAAUGCCUAUUUUUAAAGCGAUAUA (((.(((.((((((...((((...))))......(((((.(((.(((((((....))))....))).))))))))))))))...((((....)))).))))))................. (-28.56 = -29.98 + 1.42)

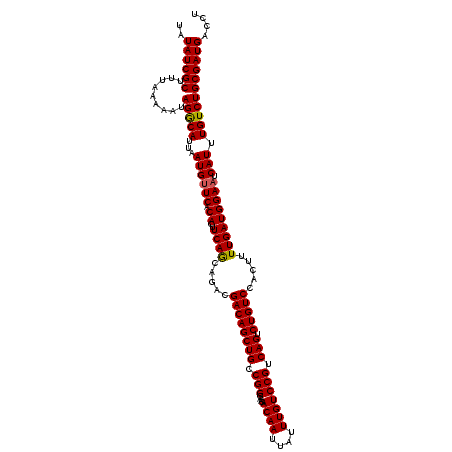
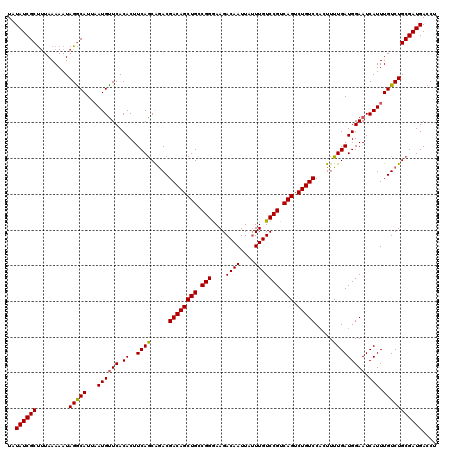
| Location | 6,979,493 – 6,979,613 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.16 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -29.56 |
| Energy contribution | -29.44 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6979493 120 - 23771897 UUUAUCGCUUUAAAAAUAGGCAUUAAUGUUCACACUUCAACAGACGACAGCUGGCGGAAAGACAAUUAUUUGUCCGUCAGUCUGUCCAAUUUUGAUGGAAUCAUUUGUCUGCGAUGACCU .(((((((.........(((((..(((((((.((..((((.....(((((((((((((.(((......))).)))))))).))))).....))))))))).))))))))))))))))... ( -36.30) >DroSec_CAF1 49369 120 - 1 UAUAUCGCUUUAAAAAUAGGCAUUAAUGUUCACACUUCAGCAGACGACAGCUGCCGGGAAGACAAUUAUUUGUCCGUCAGUCUGUCCACUUUUGAUGGAAUCAUUUGUCUGCGAUGACCU ..((((((((.......))))..................((((((((((((((.(((....((((....))))))).))).))))).......((((....)))).)))))))))).... ( -32.20) >DroSim_CAF1 49110 120 - 1 UAUAUCGCUUUAAAAAUAGGCAUUAAUGUUCACACUUCAGCAGACGACAGCUGCCGGGAAGACAAUUAUUUGUCCGUCAGUCUGUCCACUUUUGAUGGAAUCAUUUGUCUGCGAUGACCU ..((((((((.......))))..................((((((((((((((.(((....((((....))))))).))).))))).......((((....)))).)))))))))).... ( -32.20) >DroEre_CAF1 49318 119 - 1 UAUAUCGCCUUAAAAAUAGGCAU-AAUGCUCACACUUCAGCAGACGACAGCUGUCGGGAAGACAAUUACUUGUCCGUCAGUCUGUCCCAUUUUGAUGGAAUCAUUUGUCUGCGAUGACCU ..((((((((.......))))..-...............((((((((((((((.(((((((.......))).)))).))).)))))((((....))))........)))))))))).... ( -39.50) >DroYak_CAF1 50966 119 - 1 UUUAUCGCUCUAAAAAUAGACAU-UAUGUUCACACUUCAGCGAACGACAGCUGUCGGGAAGACAAUUAUUUGUCCGUCAGUCUGUCCACUUUUGAUGGAAUCAUUUGUCUGCGAUGACCU .(((((((.........(((((.-.((((((.((..((((.....((((((((.(((....((((....))))))).))).))))).....))))))))).))).))))))))))))... ( -32.80) >consensus UAUAUCGCUUUAAAAAUAGGCAUUAAUGUUCACACUUCAGCAGACGACAGCUGCCGGGAAGACAAUUAUUUGUCCGUCAGUCUGUCCACUUUUGAUGGAAUCAUUUGUCUGCGAUGACCU ..((((((.........(((((...((((((.((..((((.....((((((((.(((....((((....))))))).))).))))).....))))))))).))).))))))))))).... (-29.56 = -29.44 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:35 2006