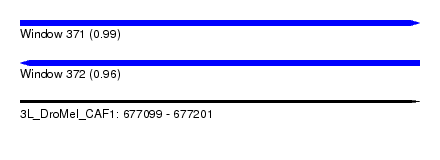
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 677,099 – 677,201 |
| Length | 102 |
| Max. P | 0.992541 |

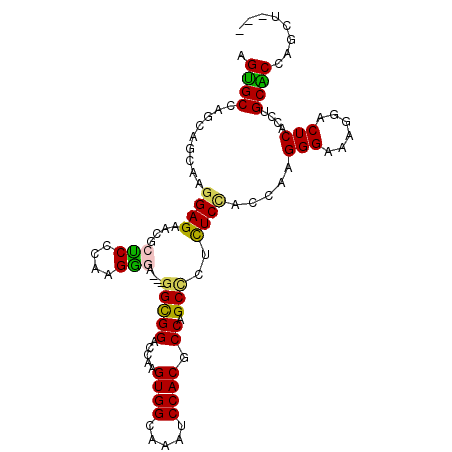
| Location | 677,099 – 677,201 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 75.79 |
| Mean single sequence MFE | -43.95 |
| Consensus MFE | -21.61 |
| Energy contribution | -21.25 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
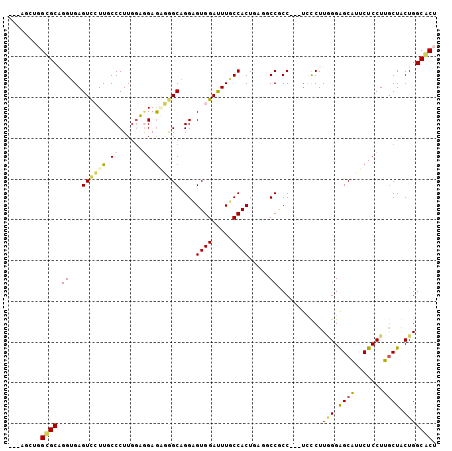
>3L_DroMel_CAF1 677099 102 + 23771897 UCGAGCUGGUGCAGGUGAGUCCUUUCCCUUGGAAGAAAGGGCAGGCGUGGAUUUGCCACUUGUUCCGCC---UCCCUUGGGGGCGUUCUCCUUGCUGCUGGCACU ....((..(((((((.((((((((((........)))))))).((((......))))........((((---(((...)))))))...))))))).))..))... ( -42.90) >DroPse_CAF1 39452 98 + 1 -------UGCGCCGGGGAGUUGUUGCCCUUGGUGGAGUUAGCUGGAGUGGAUUUUCCACUGAGGCCACCGUUUGUCUUAGGAGCAUUCUCCUUGCUACUGGCGCU -------.(((((((((((....((((((((((((..((((.(((((......)))))))))..))))))........))).)))..))))......))))))). ( -42.20) >DroSim_CAF1 32972 102 + 1 UCGAGCUGGUGCAGGUGAGUCCUUUCCCUUGGAGGAGAGGGCAGGCGUGGAUUUGCCACUUGCGCCGCC---UCCCUUGGGGGCGUUCUCCUUGCUGCUGGCACU ..((((.(((((((((..(((((((((......))))))))).((((......)))))))))))))(((---(((...))))))))))....(((.....))).. ( -51.10) >DroYak_CAF1 34780 102 + 1 UCGUGCUGGUGCAGGUGAGUCCUUUCCCUUGGAGGAGAGGGCAGGCGUGGAUUUGCCACUAGCUCCGCC---UCCCUUGGGAGCGUUCUCCUUGCUGCUGGCACU ..((((..(((((((.(((..(.(((((..(((((.(.((((.((((......))))....))))).))---)))...))))).)..)))))))).))..)))). ( -51.50) >DroAna_CAF1 34008 102 + 1 ---ACCUGGCGCAGGAGAGUCCUUGCCCUUCAUUGAGGAUGCUGGAGUGGACUUCCCACUCAGUCCGCCAUUCCCCUUUGGAGCAUUCUCCUUACUACUGGCACU ---.((.((((.(((.....))))))).......(((((((((.((((((.....))))))......(((........)))))))))))).........)).... ( -33.80) >DroPer_CAF1 43878 98 + 1 -------UGCGCCGGGGAGUUGUUGCCCUUGGUGGAGUUAGCUGGAGUGGAUUUUCCACUGAGGCCACCGUUUGUCUUAGGAGCAUUCUCCUUGCUACUGGCGCU -------.(((((((((((....((((((((((((..((((.(((((......)))))))))..))))))........))).)))..))))......))))))). ( -42.20) >consensus ___AGCUGGCGCAGGUGAGUCCUUGCCCUUGGAGGAGAGGGCAGGAGUGGAUUUGCCACUGAGGCCGCC___UCCCUUGGGAGCAUUCUCCUUGCUACUGGCACU ........((((.((...((((((.((......)).))))))....((((.....)))).....))...........(((.((((.......)))).))))))). (-21.61 = -21.25 + -0.36)

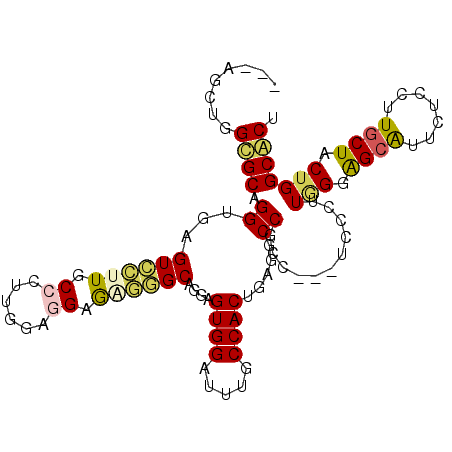
| Location | 677,099 – 677,201 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 75.79 |
| Mean single sequence MFE | -35.37 |
| Consensus MFE | -19.68 |
| Energy contribution | -18.80 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 677099 102 - 23771897 AGUGCCAGCAGCAAGGAGAACGCCCCCAAGGGA---GGCGGAACAAGUGGCAAAUCCACGCCUGCCCUUUCUUCCAAGGGAAAGGACUCACCUGCACCAGCUCGA .((((..(((((........((((((....)).---))))......((((.....))))).))))(((((((......)))))))........))))........ ( -34.20) >DroPse_CAF1 39452 98 - 1 AGCGCCAGUAGCAAGGAGAAUGCUCCUAAGACAAACGGUGGCCUCAGUGGAAAAUCCACUCCAGCUAACUCCACCAAGGGCAACAACUCCCCGGCGCA------- .(((((.((....(((((....)))))...))....(((((....((((((........)))).))....)))))..(((.........)))))))).------- ( -34.00) >DroSim_CAF1 32972 102 - 1 AGUGCCAGCAGCAAGGAGAACGCCCCCAAGGGA---GGCGGCGCAAGUGGCAAAUCCACGCCUGCCCUCUCCUCCAAGGGAAAGGACUCACCUGCACCAGCUCGA .......(.(((.((((((..(((((....)).---)))((((...((((.....))))...)))).))))))....((...(((.....)))...)).)))).. ( -35.80) >DroYak_CAF1 34780 102 - 1 AGUGCCAGCAGCAAGGAGAACGCUCCCAAGGGA---GGCGGAGCUAGUGGCAAAUCCACGCCUGCCCUCUCCUCCAAGGGAAAGGACUCACCUGCACCAGCACGA .((((..((((....(((..(..((((...(((---((.((.((..((((.....))))....))))...)))))..))))..)..)))..))))....)))).. ( -40.10) >DroAna_CAF1 34008 102 - 1 AGUGCCAGUAGUAAGGAGAAUGCUCCAAAGGGGAAUGGCGGACUGAGUGGGAAGUCCACUCCAGCAUCCUCAAUGAAGGGCAAGGACUCUCCUGCGCCAGGU--- ...(((........((((....))))...(((((...((.....((((((.....))))))..)).))))).......(((.(((.....)))..))).)))--- ( -34.10) >DroPer_CAF1 43878 98 - 1 AGCGCCAGUAGCAAGGAGAAUGCUCCUAAGACAAACGGUGGCCUCAGUGGAAAAUCCACUCCAGCUAACUCCACCAAGGGCAACAACUCCCCGGCGCA------- .(((((.((....(((((....)))))...))....(((((....((((((........)))).))....)))))..(((.........)))))))).------- ( -34.00) >consensus AGUGCCAGCAGCAAGGAGAACGCUCCCAAGGGA___GGCGGACCAAGUGGCAAAUCCACGCCAGCCCUCUCCACCAAGGGAAAGGACUCACCUGCACCAGCU___ .((((.........((((....(((....)))....(((((.....((((.....)))).)).)))..)))).....(((......)))....))))........ (-19.68 = -18.80 + -0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:26 2006