
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,976,952 – 6,977,135 |
| Length | 183 |
| Max. P | 0.984680 |

| Location | 6,976,952 – 6,977,055 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.04 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -21.13 |
| Energy contribution | -20.45 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6976952 103 + 23771897 CUUUGAACGCGUGCGCAACGCAAGCCGUUUAAUCUGAAUUUGCUCACUCAAUGUGGGCUGGAAUAUAUAU-------UAUAUGUAUA----------UAUAUAUUCAUUUUCGGCCACUA ..((((((((.((((...)))).).))))))).(((((...((((((.....))))))..((((((((((-------.........)----------)))))))))...)))))...... ( -29.30) >DroSim_CAF1 46638 110 + 1 CUUUGAACGCGUGCGCAAUGCAACCCGUUUAAUCUAAAUUUGUUCACUCAAUGUGGGCUGGAAUAUAUAUAUAUAUUGAUUUAUAUG----------UAUAUAUUCAUUUUCGGCCACUA ..(((((((..(((.....)))...)))))))............(((.....)))((((((((((((((((((((......))))))----------))))))))).....))))).... ( -31.50) >DroEre_CAF1 46798 112 + 1 CUUUGAACGCGUGCGCAACGCAACCUGUUUAAUCUGAAUUUGCUCACCCAGUGUGGGCUGGAAUAAAUAUG--------AGUAUGCACACAUACGGGUAUGUCUUUCUUUUCGGCCACUA ..(((((((..((((...))))...))))))).(((((...((((((.....)))))).((((...((((.--------.(((((....)))))..))))....)))).)))))...... ( -26.80) >DroYak_CAF1 48284 102 + 1 CUUUGAACGCGUGCGCAACGCAACCUGUUUAAUCUGAAUUUGCUCACCCAAUGUGGGCUGGAAUACAUAUA--------UUUAUAUA----------UAUAUGUUCAUUUUCGGCUACUA ..(((((((..((((...))))...))))))).(((((...((((((.....))))))..(((((.(((((--------(....)))----------))).)))))...)))))...... ( -25.10) >consensus CUUUGAACGCGUGCGCAACGCAACCCGUUUAAUCUGAAUUUGCUCACCCAAUGUGGGCUGGAAUAUAUAUA________UUUAUAUA__________UAUAUAUUCAUUUUCGGCCACUA ..(((((((..((((...))))...)))))))............(((.....)))((((((((.(((((((..........................)))))))....)))))))).... (-21.13 = -20.45 + -0.69)



| Location | 6,976,992 – 6,977,095 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.54 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -19.06 |
| Energy contribution | -18.62 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6976992 103 + 23771897 UGCUCACUCAAUGUGGGCUGGAAUAUAUAU-------UAUAUGUAUA----------UAUAUAUUCAUUUUCGGCCACUAUCUGAUCGAUCGAAAGACAAUUAGCCAAAUAAGUCGACGG .((((((.....))))))..((((((((((-------.........)----------)))))))))....(((((......(((((.(.((....))).)))))........)))))... ( -26.54) >DroSim_CAF1 46678 110 + 1 UGUUCACUCAAUGUGGGCUGGAAUAUAUAUAUAUAUUGAUUUAUAUG----------UAUAUAUUCAUUUUCGGCCACUAUCUGAUCGAUCGAAAGACAAUUAGCCAAAUAAGUCGACGG .((((((.....))))))..(((((((((((((((......))))))----------)))))))))....(((((......(((((.(.((....))).)))))........)))))... ( -31.44) >DroEre_CAF1 46838 112 + 1 UGCUCACCCAGUGUGGGCUGGAAUAAAUAUG--------AGUAUGCACACAUACGGGUAUGUCUUUCUUUUCGGCCACUAUCUGAUCGAUCGAAAGACAAUUAGCCAAAUAAGUCGACGG .((((((.....))))))(((.....((((.--------.(((((....)))))..))))(((((((...(((..((.....))..)))..)))))))......)))............. ( -29.70) >DroYak_CAF1 48324 102 + 1 UGCUCACCCAAUGUGGGCUGGAAUACAUAUA--------UUUAUAUA----------UAUAUGUUCAUUUUCGGCUACUAUCCGAUCGAUCGAAAGACAAUUAGCCAAAUAAGUCGACGG ....(((.....)))((((((((.(((((((--------(......)----------)))))))....)))))))).....(((.((((((....))...(((......))).))))))) ( -24.40) >consensus UGCUCACCCAAUGUGGGCUGGAAUAUAUAUA________UUUAUAUA__________UAUAUAUUCAUUUUCGGCCACUAUCUGAUCGAUCGAAAGACAAUUAGCCAAAUAAGUCGACGG ....(((.....)))((((((((.(((((((..........................)))))))....)))))))).....(((.((((((....))...(((......))).))))))) (-19.06 = -18.62 + -0.44)

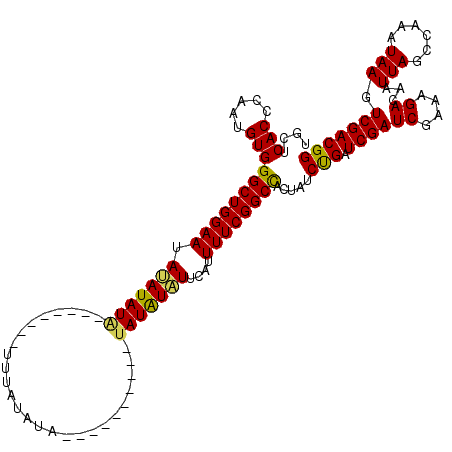
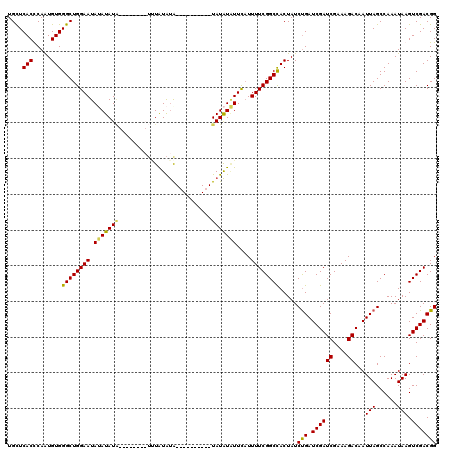
| Location | 6,976,992 – 6,977,095 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.54 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -20.30 |
| Energy contribution | -20.36 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6976992 103 - 23771897 CCGUCGACUUAUUUGGCUAAUUGUCUUUCGAUCGAUCAGAUAGUGGCCGAAAAUGAAUAUAUA----------UAUACAUAUA-------AUAUAUAUUCCAGCCCACAUUGAGUGAGCA ...........((((((((.((((((.((....))..))))))))))))))...(((((((((----------(.........-------))))))))))..((.(((.....))).)). ( -26.40) >DroSim_CAF1 46678 110 - 1 CCGUCGACUUAUUUGGCUAAUUGUCUUUCGAUCGAUCAGAUAGUGGCCGAAAAUGAAUAUAUA----------CAUAUAAAUCAAUAUAUAUAUAUAUUCCAGCCCACAUUGAGUGAACA ..((..(((((((((((((.((((((.((....))..))))))))))))))...(((((((((----------.(((((......))))).)))))))))..........)))))..)). ( -28.80) >DroEre_CAF1 46838 112 - 1 CCGUCGACUUAUUUGGCUAAUUGUCUUUCGAUCGAUCAGAUAGUGGCCGAAAAGAAAGACAUACCCGUAUGUGUGCAUACU--------CAUAUUUAUUCCAGCCCACACUGGGUGAGCA ..(((..(((.((((((((.((((((.((....))..)))))))))))))))))...))).((((((..((((.((.....--------.............)).)))).)))))).... ( -31.07) >DroYak_CAF1 48324 102 - 1 CCGUCGACUUAUUUGGCUAAUUGUCUUUCGAUCGAUCGGAUAGUAGCCGAAAAUGAACAUAUA----------UAUAUAAA--------UAUAUGUAUUCCAGCCCACAUUGGGUGAGCA .....(.(((.((((((((.((((((.((....))..)))))))))))))).....(((((((----------(......)--------)))))))......((((.....)))))))). ( -27.00) >consensus CCGUCGACUUAUUUGGCUAAUUGUCUUUCGAUCGAUCAGAUAGUGGCCGAAAAUGAAUAUAUA__________UAUAUAAA________UAUAUAUAUUCCAGCCCACAUUGAGUGAGCA ..((..(((((((((((((.((((((.((....))..))))))))))))))...(((((((((............................)))))))))..........)))))..)). (-20.30 = -20.36 + 0.06)

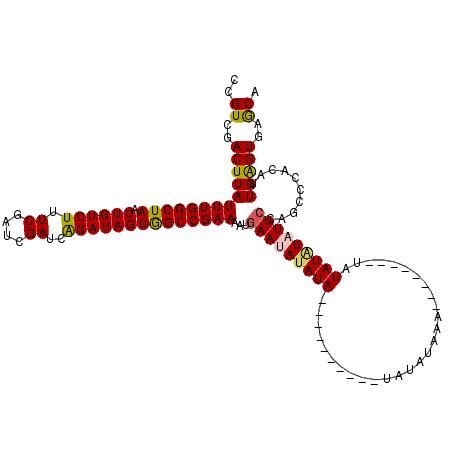

| Location | 6,977,025 – 6,977,135 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.96 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -27.96 |
| Energy contribution | -27.02 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6977025 110 - 23771897 AUUAAUUAAGACGCCGGCACGGGUACGAUCUCUCUGCUGCCCGUCGACUUAUUUGGCUAAUUGUCUUUCGAUCGAUCAGAUAGUGGCCGAAAAUGAAUAUAUA----------UAUACAU .........((((.(((((.(((........))))))))..))))......((((((((.((((((.((....))..))))))))))))))............----------....... ( -27.90) >DroSim_CAF1 46718 110 - 1 AUUAAUUAAGACGCCGGCACGGGUACGAUCUCUCUGCUGCCCGUCGACUUAUUUGGCUAAUUGUCUUUCGAUCGAUCAGAUAGUGGCCGAAAAUGAAUAUAUA----------CAUAUAA .........((((.(((((.(((........))))))))..))))......((((((((.((((((.((....))..))))))))))))))............----------....... ( -27.90) >DroEre_CAF1 46870 120 - 1 AUUAAUUAAGACGCCGGCACAGGUCGGAUCUCUCUGCUGCCCGUCGACUUAUUUGGCUAAUUGUCUUUCGAUCGAUCAGAUAGUGGCCGAAAAGAAAGACAUACCCGUAUGUGUGCAUAC .........((((..((((((((.........)))).)))))))).........((.....((((((((..(((..((.....))..)))...))))))))...))(((((....))))) ( -32.40) >DroYak_CAF1 48356 110 - 1 AUUAAUUGAGACGCGGGCACGGGUUCGAUCUCUCCGCUGUCCGUCGACUUAUUUGGCUAAUUGUCUUUCGAUCGAUCGGAUAGUAGCCGAAAAUGAACAUAUA----------UAUAUAA ......((((.((((((((((((.........)))).)))))).)).))))((((((((.((((((.((....))..))))))))))))))............----------....... ( -32.20) >consensus AUUAAUUAAGACGCCGGCACGGGUACGAUCUCUCUGCUGCCCGUCGACUUAUUUGGCUAAUUGUCUUUCGAUCGAUCAGAUAGUGGCCGAAAAUGAAUAUAUA__________UAUAUAA .........((((..((((((((.........)))).))))))))......((((((((.((((((.((....))..))))))))))))))............................. (-27.96 = -27.02 + -0.94)

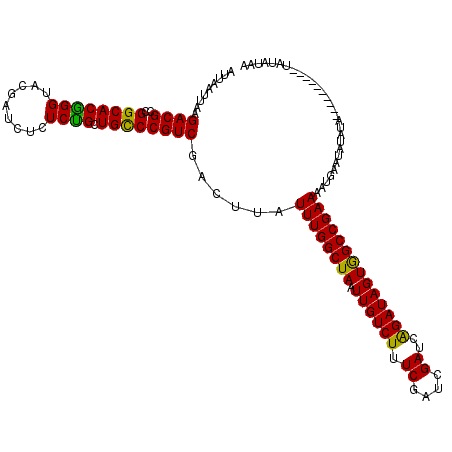

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:29 2006