
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,976,498 – 6,976,792 |
| Length | 294 |
| Max. P | 0.978353 |

| Location | 6,976,498 – 6,976,613 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.57 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -17.80 |
| Energy contribution | -18.72 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.680961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6976498 115 + 23771897 AAUAUUGCAUUUACAUUUUGUGCAUUCUCGGAAUAAAAUCGCCUGCAAUUUCAGUGAAUCAACAUUAAUUCGCUGACCUUGCCAUUAAAUAUUUUAUCGUUGCUUGUGUUCCGCU----- .....(((((.........)))))....(((((((.........((((..(((((((((........)))))))))..))))((...(((........)))...)))))))))..----- ( -24.40) >DroSec_CAF1 46398 115 + 1 AAUAUUGCAUAUAUUUUUUGUGCAUUCCCGGAAUAAAAUCGCCUGCAAUCUCAGUGAAUCAACAUUAUUCCGCUGACCUUGCCAUUAAAUAUUUGAUCGUUGCUUGUGUUCCGCU----- .....((((((.......))))))....(((((((.....((..((((..(((((((((.......))).))))))..)))).(((((....)))))....))...)))))))..----- ( -23.40) >DroSim_CAF1 46184 115 + 1 AAUAUUGCAUUUAUUUUUUGUGCAUUCCCGGAAUAAAAUCGCCUGCAAUCUCAGUGAAUCAACAUUAUUUCGCUGACCUUGCCAUUAAAUAUUUGAUCGUUGCUUGUGUUCCGCU----- .....(((((.........)))))....(((((((.....((..((((..((((((((..........))))))))..)))).(((((....)))))....))...)))))))..----- ( -24.80) >DroEre_CAF1 46353 110 + 1 UGUAUUGCACU-----UUUGCACAUACACGGAAUACAAUUUCCUGCAAUUUCGGUGAAUCAAAAUUAUUUCGCUGACCUUGCCAUUAAAUAUUUUAUCGUGCCUUGUUUUCCGCU----- (((((((((..-----..)))).)))))(((((.((((......((((..((((((((..........))))))))..))))(((.............)))..)))).)))))..----- ( -25.42) >DroYak_CAF1 47825 119 + 1 UAUAUUGCAUUUAUU-UGUGUGUAUUCCCGGAAUAAAAUCUGCUGCAAUUUCAGUAAAUCAACAUUAUUUCGCUGACCUUGCCAUUAAAUAUUUUAUCGUUGCUUGUUUUCCGCUUGUGU .....(((((.....-...)))))....(((((...(((..((.((((..((((..((..........))..))))..)))).....(((........)))))..))))))))....... ( -15.50) >consensus AAUAUUGCAUUUAUUUUUUGUGCAUUCCCGGAAUAAAAUCGCCUGCAAUUUCAGUGAAUCAACAUUAUUUCGCUGACCUUGCCAUUAAAUAUUUUAUCGUUGCUUGUGUUCCGCU_____ .....(((((.........)))))....(((((((.........((((..((((((((..........))))))))..))))((...(((........)))...)))))))))....... (-17.80 = -18.72 + 0.92)



| Location | 6,976,538 – 6,976,640 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.45 |
| Mean single sequence MFE | -19.15 |
| Consensus MFE | -15.15 |
| Energy contribution | -15.43 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6976538 102 - 23771897 AACAAUGACAACAAGUUCCGAAAAAA-A-----------------AGCGGAACACAAGCAACGAUAAAAUAUUUAAUGGCAAGGUCAGCGAAUUAAUGUUGAUUCACUGAAAUUGCAGGC ..............((((((......-.-----------------..)))))).........................((((..((((.((((((....)))))).))))..)))).... ( -24.20) >DroSec_CAF1 46438 103 - 1 AACAAUGACAACAAGUUCCGAAAAAAUA-----------------AGCGGAACACAAGCAACGAUCAAAUAUUUAAUGGCAAGGUCAGCGGAAUAAUGUUGAUUCACUGAGAUUGCAGGC ..............((((((........-----------------..)))))).........................((((..((((..((((.......)))).))))..)))).... ( -19.30) >DroSim_CAF1 46224 103 - 1 AACAAUGACAACAAGUUCCGAAAAAAUA-----------------AGCGGAACACAAGCAACGAUCAAAUAUUUAAUGGCAAGGUCAGCGAAAUAAUGUUGAUUCACUGAGAUUGCAGGC ..............((((((........-----------------..)))))).........................((((..((((.(((.((....)).))).))))..)))).... ( -19.50) >DroEre_CAF1 46388 99 - 1 AACAAUGAC---AAGUUCCGAAAAAA-A-----------------AGCGGAAAACAAGGCACGAUAAAAUAUUUAAUGGCAAGGUCAGCGAAAUAAUUUUGAUUCACCGAAAUUGCAGGA .....((((---...(((((......-.-----------------..)))))......((.((.(((.....))).))))...))))......((((((((......))))))))..... ( -13.90) >DroYak_CAF1 47864 116 - 1 AACAAUGAC---AAGUUCCGAAAAAA-AAAAAAAAACGGAACACAAGCGGAAAACAAGCAACGAUAAAAUAUUUAAUGGCAAGGUCAGCGAAAUAAUGUUGAUUUACUGAAAUUGCAGCA .........---..((((((......-.........))))))....(((.....).......................((((..((((..((.((....)).))..))))..)))).)). ( -18.86) >consensus AACAAUGACAACAAGUUCCGAAAAAA_A_________________AGCGGAACACAAGCAACGAUAAAAUAUUUAAUGGCAAGGUCAGCGAAAUAAUGUUGAUUCACUGAAAUUGCAGGC ..............((((((...........................)))))).........................((((..((((.(((.((....)).))).))))..)))).... (-15.15 = -15.43 + 0.28)



| Location | 6,976,613 – 6,976,715 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.62 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -16.89 |
| Energy contribution | -17.25 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6976613 102 + 23771897 ------------U-UUUUUUCGGAACUUGUUGUCAUUGUUUA-----UAAAUGCAGGACAAUGGCCAUUGUCUCCUGUUGUUUAUGCCUACGAAUUGAAAAUACAUAAACAAAGACAUUG ------------.-.......((...(..(((((.((((...-----.....)))))))))..)))..(((((....(((((((((.................))))))))))))))... ( -20.43) >DroSec_CAF1 46513 103 + 1 ------------UAUUUUUUCGGAACUUGUUGUCAUUGUUUA-----UAAAUGCAGGACAAUGGCCAUUGUCUCCUGUUGUUUAUGCCUUCGAAUUGGAAAUGUAUAAACAAAGACAUUG ------------.........((...(..(((((.((((...-----.....)))))))))..)))..(((((....((((((((((.(((......)))..)))))))))))))))... ( -24.30) >DroSim_CAF1 46299 103 + 1 ------------UAUUUUUUCGGAACUUGUUGUCAUUGUUUA-----UAAAUGCAGGACAAUGGCCAUUGUCUCCUGUUGUUUAUGCCUUCGAAUUGGAAAUGCAUAAACAAAGACAUUG ------------.........((...(..(((((.((((...-----.....)))))))))..)))..(((((....((((((((((.(((......)))..)))))))))))))))... ( -26.30) >DroEre_CAF1 46463 104 + 1 ------------U-UUUUUUCGGAACUU---GUCAUUGUUUGUAUGGUAAAUGCAGGACAAUGGCCAUUGUGUCCAGUUGUUUAUGCAUUCGAAUUGGUAAUACGUAAACAAAGACAUUG ------------.-..............---((((((((((((((.....))))).)))))))))....(((((...(((((((((.((((......).))).))))))))).))))).. ( -26.20) >DroYak_CAF1 47944 107 + 1 UCCGUUUUUUUUU-UUUUUUCGGAACUU---GUCAUUGUUUA-----UAAAUGCAGGACAAUGGCCAUUGUGUCCUGUUGUUUAUGCAUUCGCAUUGCAAAU----AAACAAAGACAUUG ((((.........-......))))...(---(((.(((((((-----(....((((((((..........))))))))(((..((((....)))).))).))----)))))).))))... ( -29.36) >consensus ____________U_UUUUUUCGGAACUUGUUGUCAUUGUUUA_____UAAAUGCAGGACAAUGGCCAUUGUCUCCUGUUGUUUAUGCCUUCGAAUUGGAAAUACAUAAACAAAGACAUUG .....................(((((.....((((((((((..............))))))))))....)).)))..(((((((((.................)))))))))........ (-16.89 = -17.25 + 0.36)

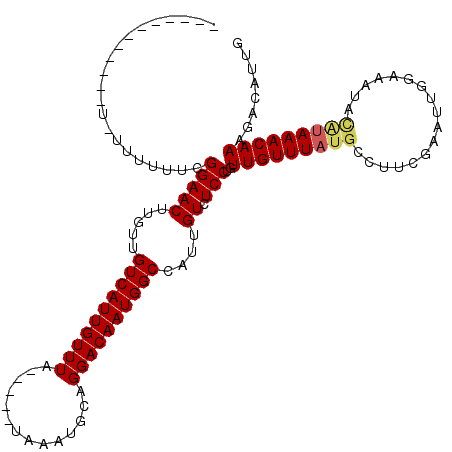

| Location | 6,976,613 – 6,976,715 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.62 |
| Mean single sequence MFE | -21.45 |
| Consensus MFE | -12.80 |
| Energy contribution | -14.44 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6976613 102 - 23771897 CAAUGUCUUUGUUUAUGUAUUUUCAAUUCGUAGGCAUAAACAACAGGAGACAAUGGCCAUUGUCCUGCAUUUA-----UAAACAAUGACAACAAGUUCCGAAAAAA-A------------ ...((((.((((((((((.....(.....)...))))))))))(((((..((((....)))))))))......-----........))))................-.------------ ( -19.00) >DroSec_CAF1 46513 103 - 1 CAAUGUCUUUGUUUAUACAUUUCCAAUUCGAAGGCAUAAACAACAGGAGACAAUGGCCAUUGUCCUGCAUUUA-----UAAACAAUGACAACAAGUUCCGAAAAAAUA------------ ...((((.(((((((((..((((......))))..........(((((..((((....)))))))))....))-----))))))).))))..................------------ ( -19.70) >DroSim_CAF1 46299 103 - 1 CAAUGUCUUUGUUUAUGCAUUUCCAAUUCGAAGGCAUAAACAACAGGAGACAAUGGCCAUUGUCCUGCAUUUA-----UAAACAAUGACAACAAGUUCCGAAAAAAUA------------ ...((((.((((((((((.((((......))))))))))))))(((((..((((....)))))))))......-----........))))..................------------ ( -23.90) >DroEre_CAF1 46463 104 - 1 CAAUGUCUUUGUUUACGUAUUACCAAUUCGAAUGCAUAAACAACUGGACACAAUGGCCAUUGUCCUGCAUUUACCAUACAAACAAUGAC---AAGUUCCGAAAAAA-A------------ ...((((.((((((..((((.........(((((((...((((.(((.(.....).)))))))..)))))))...)))))))))).)))---).............-.------------ ( -18.30) >DroYak_CAF1 47944 107 - 1 CAAUGUCUUUGUUU----AUUUGCAAUGCGAAUGCAUAAACAACAGGACACAAUGGCCAUUGUCCUGCAUUUA-----UAAACAAUGAC---AAGUUCCGAAAAAA-AAAAAAAAACGGA ...((((.((((((----((.(((.((((....)))).......((((((..........)))))))))...)-----))))))).)))---)...((((......-.........)))) ( -26.36) >consensus CAAUGUCUUUGUUUAUGCAUUUCCAAUUCGAAGGCAUAAACAACAGGAGACAAUGGCCAUUGUCCUGCAUUUA_____UAAACAAUGACAACAAGUUCCGAAAAAA_A____________ ....(((.((((((((((.(((.......))).))))))))))(((((..((((....)))))))))...................)))............................... (-12.80 = -14.44 + 1.64)

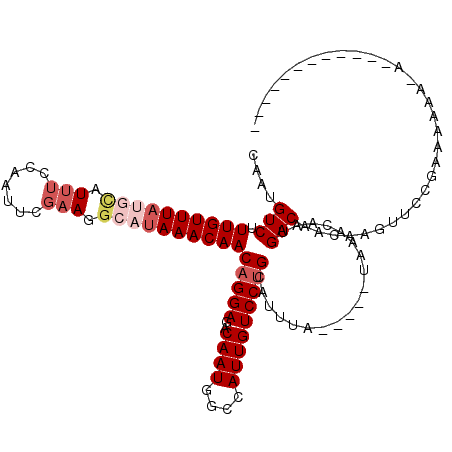

| Location | 6,976,640 – 6,976,753 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.89 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -20.14 |
| Energy contribution | -19.34 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6976640 113 + 23771897 UA-----UAAAUGCAGGACAAUGGCCAUUGUCUCCUGUUGUUUAUGCCUACGAAUUGAAAAUACAUAAACAAAGACAUUGUCUUUGAUCAAAAUUCUCAGUG--AUUGUAGUGCACUUUC .(-----(((((((((((((((....))))..))))).)))))))(((((((((((((.(((.......(((((((...)))))))......))).))))).--.)))))).))...... ( -27.52) >DroSec_CAF1 46541 115 + 1 UA-----UAAAUGCAGGACAAUGGCCAUUGUCUCCUGUUGUUUAUGCCUUCGAAUUGGAAAUGUAUAAACAAAGACAUUGUCUUUGAUCAUAAUUUCCAGUGAGACUGCACUGCACUUUC ..-----....(((((((((((....))))))............(((..((..(((((((((.......(((((((...)))))))......)))))))))..))..))))))))..... ( -33.82) >DroSim_CAF1 46327 115 + 1 UA-----UAAAUGCAGGACAAUGGCCAUUGUCUCCUGUUGUUUAUGCCUUCGAAUUGGAAAUGCAUAAACAAAGACAUUGUCUUUGAUCAUAAUUCGCAGUGAGACUGCACUGCACUUUC ..-----....((((((((((.(((...(((((....((((((((((.(((......)))..)))))))))))))))..))).))).)).......((((.....)))).)))))..... ( -34.80) >DroEre_CAF1 46487 118 + 1 UGUAUGGUAAAUGCAGGACAAUGGCCAUUGUGUCCAGUUGUUUAUGCAUUCGAAUUGGUAAUACGUAAACAAAGACAUUGUCUUUGAUCAUAAUUCCUAGUG--ACUGUACUAUACUUUC .((((((((..(((((((..(((..((..(((((...(((((((((.((((......).))).))))))))).)))))......))..)))...)))).)))--....)))))))).... ( -28.90) >DroYak_CAF1 47980 109 + 1 UA-----UAAAUGCAGGACAAUGGCCAUUGUGUCCUGUUGUUUAUGCAUUCGCAUUGCAAAU----AAACAAAGACAUUGUCUUUGAUCAUAAUUCGCAGUG--ACUGUACUACACUUUC .(-----(((((((((((((..........))))))).)))))))(((....((((((.(((----...(((((((...)))))))......))).))))))--..)))........... ( -25.60) >consensus UA_____UAAAUGCAGGACAAUGGCCAUUGUCUCCUGUUGUUUAUGCCUUCGAAUUGGAAAUACAUAAACAAAGACAUUGUCUUUGAUCAUAAUUCCCAGUG__ACUGUACUGCACUUUC ............((((((((((....))))..))))))(((...(((......(((((.(((.......(((((((...)))))))......))).)))))......)))..)))..... (-20.14 = -19.34 + -0.80)

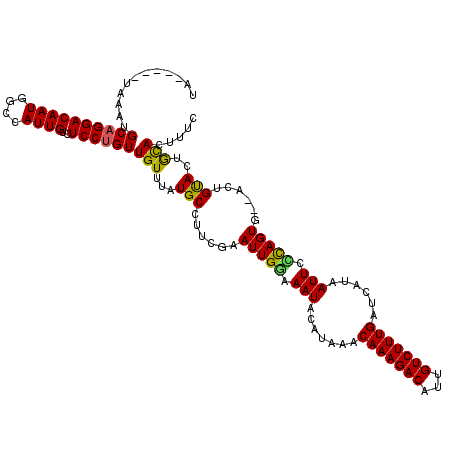

| Location | 6,976,640 – 6,976,753 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.89 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -22.38 |
| Energy contribution | -22.42 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6976640 113 - 23771897 GAAAGUGCACUACAAU--CACUGAGAAUUUUGAUCAAAGACAAUGUCUUUGUUUAUGUAUUUUCAAUUCGUAGGCAUAAACAACAGGAGACAAUGGCCAUUGUCCUGCAUUUA-----UA ..((((((.(((((((--...(((((((..(((.(((((((...))))))).)))...)))))))))).))))............((.((((((....)))))))))))))).-----.. ( -28.20) >DroSec_CAF1 46541 115 - 1 GAAAGUGCAGUGCAGUCUCACUGGAAAUUAUGAUCAAAGACAAUGUCUUUGUUUAUACAUUUCCAAUUCGAAGGCAUAAACAACAGGAGACAAUGGCCAUUGUCCUGCAUUUA-----UA ..(((((((((((..((....((((((((((((.(((((((...))))))).))))).)))))))....))..)))............((((((....)))))))))))))).-----.. ( -36.50) >DroSim_CAF1 46327 115 - 1 GAAAGUGCAGUGCAGUCUCACUGCGAAUUAUGAUCAAAGACAAUGUCUUUGUUUAUGCAUUUCCAAUUCGAAGGCAUAAACAACAGGAGACAAUGGCCAUUGUCCUGCAUUUA-----UA ..(((((((((((((.....))))).............(((((((.((((((((((((.((((......))))))))))))))..(....)...)).))))))))))))))).-----.. ( -38.40) >DroEre_CAF1 46487 118 - 1 GAAAGUAUAGUACAGU--CACUAGGAAUUAUGAUCAAAGACAAUGUCUUUGUUUACGUAUUACCAAUUCGAAUGCAUAAACAACUGGACACAAUGGCCAUUGUCCUGCAUUUACCAUACA ....((((.((.....--.....((...((((..(((((((...)))))))....))))...)).....(((((((...((((.(((.(.....).)))))))..))))))))).)))). ( -24.70) >DroYak_CAF1 47980 109 - 1 GAAAGUGUAGUACAGU--CACUGCGAAUUAUGAUCAAAGACAAUGUCUUUGUUU----AUUUGCAAUGCGAAUGCAUAAACAACAGGACACAAUGGCCAUUGUCCUGCAUUUA-----UA ..((((((((....((--(..(((((((...((.(((((((...))))))))).----)))))))((((....)))).........)))(((((....))))).)))))))).-----.. ( -27.90) >consensus GAAAGUGCAGUACAGU__CACUGCGAAUUAUGAUCAAAGACAAUGUCUUUGUUUAUGCAUUUCCAAUUCGAAGGCAUAAACAACAGGAGACAAUGGCCAUUGUCCUGCAUUUA_____UA ..((((((((.(((((.....(((((((..(((.(((((((...))))))).)))...)))))))....................(....).......))))).))))))))........ (-22.38 = -22.42 + 0.04)



| Location | 6,976,675 – 6,976,792 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.92 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -17.34 |
| Energy contribution | -17.98 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6976675 117 - 23771897 AUUAAAGCGGGA-AAUAUCCAAAUGUCUUUCGCAGGUGACGAAAGUGCACUACAAU--CACUGAGAAUUUUGAUCAAAGACAAUGUCUUUGUUUAUGUAUUUUCAAUUCGUAGGCAUAAA ......(((..(-.((((....)))).)..))).(....)....((((.(((((((--...(((((((..(((.(((((((...))))))).)))...)))))))))).))))))))... ( -30.20) >DroSec_CAF1 46576 119 - 1 AUUAACGAGGAA-AAUAUCUAAAUGUUUUCCACAGGUGACGAAAGUGCAGUGCAGUCUCACUGGAAAUUAUGAUCAAAGACAAUGUCUUUGUUUAUACAUUUCCAAUUCGAAGGCAUAAA ........((((-(((((....)))))))))....((.((....)))).((((..((....((((((((((((.(((((((...))))))).))))).)))))))....))..))))... ( -35.00) >DroSim_CAF1 46362 119 - 1 AUUAAAGAGGAA-AAUAUAUAAAUGUUUUCCACAGGUGACGAAAGUGCAGUGCAGUCUCACUGCGAAUUAUGAUCAAAGACAAUGUCUUUGUUUAUGCAUUUCCAAUUCGAAGGCAUAAA ........((((-(((((....))))))))).......((....))((((((......))))))..........(((((((...)))))))(((((((.((((......))))))))))) ( -32.50) >DroEre_CAF1 46527 115 - 1 AUUAAAGGGG---AUUAUCUAAAUGCUUUUCGCAGGUGACGAAAGUAUAGUACAGU--CACUAGGAAUUAUGAUCAAAGACAAUGUCUUUGUUUACGUAUUACCAAUUCGAAUGCAUAAA ..........---.........((((..((((..((((((..............))--)))).((...((((..(((((((...)))))))....))))...))....)))).))))... ( -23.44) >DroYak_CAF1 48015 114 - 1 AUGAAAGGGGGUAAUCAUCUAAAUGGUUUUUGUAGGUGACGAAAGUGUAGUACAGU--CACUGCGAAUUAUGAUCAAAGACAAUGUCUUUGUUU----AUUUGCAAUGCGAAUGCAUAAA ........((((....)))).....((.((((((((((((..............))--))))((((((...((.(((((((...))))))))).----))))))..)))))).))..... ( -24.44) >consensus AUUAAAGAGGAA_AAUAUCUAAAUGUUUUUCGCAGGUGACGAAAGUGCAGUACAGU__CACUGCGAAUUAUGAUCAAAGACAAUGUCUUUGUUUAUGCAUUUCCAAUUCGAAGGCAUAAA ......................((((((((........((....))...............(((((((..(((.(((((((...))))))).)))...)))))))....))))))))... (-17.34 = -17.98 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:26 2006