
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,975,099 – 6,975,327 |
| Length | 228 |
| Max. P | 0.957111 |

| Location | 6,975,099 – 6,975,218 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.98 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -25.57 |
| Energy contribution | -25.93 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6975099 119 + 23771897 UAAUUAAAUGACAAAUGCCCCGAAUUUGGCAGGGAAAAAAGUUGUGACAGCAAACAUUUUUGCCGCACAUUUGUGUUAACAUUCGUGGGCUUGACUGUUGAAAGUUUCCCCUUUUGUUG- .........((((((((((........))))((((((...((((...)))).((((.((..(((.(((...(((....)))...))))))..)).)))).....))))))..)))))).- ( -30.80) >DroSec_CAF1 45006 119 + 1 UAAUUAAAUGACAAAUGCCCCGAAUUUGGCAGGGAAAAAAGUUGUGACAGCAAACAUUUUUGCCACACAUUUGUGUUAACAUUCGUGGGCUUGACUGUUGAAAGUUUCCCCUUUUGUUG- .........((((((((((........))))((((((...((((...)))).((((.((..(((.(((...(((....)))...))))))..)).)))).....))))))..)))))).- ( -30.80) >DroSim_CAF1 44796 119 + 1 UAAUUAAAUGACAAAUGCCCCGAAUUUGGCAGGGAAAAAAGUUGUGACAGCAAACAUUUUUGCCACACAUUUGUGUUAACAUUCGUGGGCUUGACUGUUGAAAGUUUCCCCUUUUGUUG- .........((((((((((........))))((((((...((((...)))).((((.((..(((.(((...(((....)))...))))))..)).)))).....))))))..)))))).- ( -30.80) >DroEre_CAF1 44941 119 + 1 UAAUUAAAUGACAAAUGCCCCACAUUUGGCAGGGAAAAAAGUUGUGACAGCAAACAUUUUUGGCACACAUUUGUGUUAACAUUUGUGGGCUUGACUGUUGAAAGUUCCCCCUUUUGUUG- .........((((((((((........))))(((((........(((((((((.((....))((.((((..(((....)))..)))).))))).))))))....)))))...)))))).- ( -30.40) >DroYak_CAF1 46277 120 + 1 UAAUUAAAUGACAAAUGCCCCGAAUUUGGCUGGAAAAAAAGUUGUGACAGCAAACAUUUUUGGCACACAUUUGUGCUAACAUUCGUGGGCUUGACAGUUGAAAGUUCCCCCUUUUGUUGC .........((((((.(((((((((...((((...............))))........((((((((....)))))))).))))).))))..(((........)))......)))))).. ( -28.86) >consensus UAAUUAAAUGACAAAUGCCCCGAAUUUGGCAGGGAAAAAAGUUGUGACAGCAAACAUUUUUGCCACACAUUUGUGUUAACAUUCGUGGGCUUGACUGUUGAAAGUUUCCCCUUUUGUUG_ .........((((((.(((((((((((((((.(((.......((((...(((((....)))))..))))))).)))))).))))).))))..((((......))))......)))))).. (-25.57 = -25.93 + 0.36)

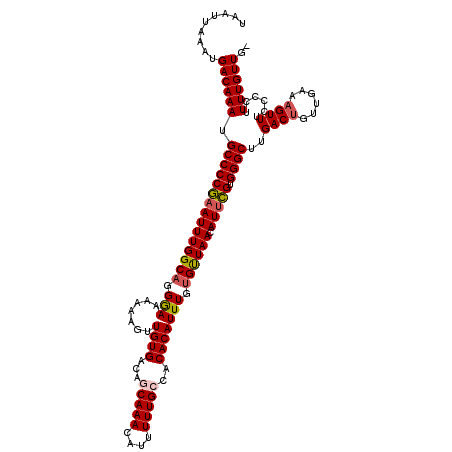

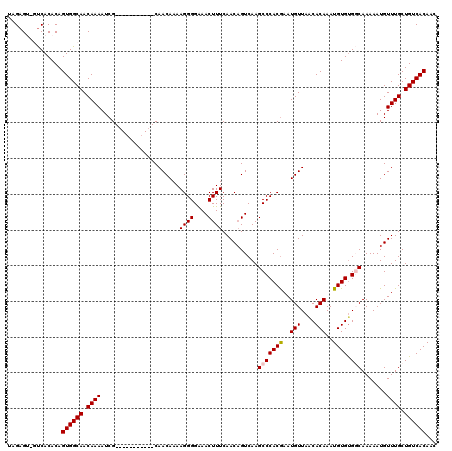
| Location | 6,975,099 – 6,975,218 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.98 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -23.78 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6975099 119 - 23771897 -CAACAAAAGGGGAAACUUUCAACAGUCAAGCCCACGAAUGUUAACACAAAUGUGCGGCAAAAAUGUUUGCUGUCACAACUUUUUUCCCUGCCAAAUUCGGGGCAUUUGUCAUUUAAUUA -..(((((((((((((.....((((.((........)).))))........(((((((((((....))))))).))))....))))))))(((........))).))))).......... ( -30.00) >DroSec_CAF1 45006 119 - 1 -CAACAAAAGGGGAAACUUUCAACAGUCAAGCCCACGAAUGUUAACACAAAUGUGUGGCAAAAAUGUUUGCUGUCACAACUUUUUUCCCUGCCAAAUUCGGGGCAUUUGUCAUUUAAUUA -..(((((((((((((......((((.(((((.......(((((.(((....)))))))).....)))))))))........))))))))(((........))).))))).......... ( -27.44) >DroSim_CAF1 44796 119 - 1 -CAACAAAAGGGGAAACUUUCAACAGUCAAGCCCACGAAUGUUAACACAAAUGUGUGGCAAAAAUGUUUGCUGUCACAACUUUUUUCCCUGCCAAAUUCGGGGCAUUUGUCAUUUAAUUA -..(((((((((((((......((((.(((((.......(((((.(((....)))))))).....)))))))))........))))))))(((........))).))))).......... ( -27.44) >DroEre_CAF1 44941 119 - 1 -CAACAAAAGGGGGAACUUUCAACAGUCAAGCCCACAAAUGUUAACACAAAUGUGUGCCAAAAAUGUUUGCUGUCACAACUUUUUUCCCUGCCAAAUGUGGGGCAUUUGUCAUUUAAUUA -..(((((((((..((......((((.(((((.......((...((((....))))..)).....)))))))))........))..))))(((........))).))))).......... ( -25.14) >DroYak_CAF1 46277 120 - 1 GCAACAAAAGGGGGAACUUUCAACUGUCAAGCCCACGAAUGUUAGCACAAAUGUGUGCCAAAAAUGUUUGCUGUCACAACUUUUUUUCCAGCCAAAUUCGGGGCAUUUGUCAUUUAAUUA ...(((((((((....))))..........((((.(((((....(((((....)))))...........((((...............))))...))))))))).))))).......... ( -25.46) >consensus _CAACAAAAGGGGAAACUUUCAACAGUCAAGCCCACGAAUGUUAACACAAAUGUGUGGCAAAAAUGUUUGCUGUCACAACUUUUUUCCCUGCCAAAUUCGGGGCAUUUGUCAUUUAAUUA ...(((((((((....))))..........((((.(((((((....))...(((((((((((....))))))).)))).................))))))))).))))).......... (-23.78 = -23.90 + 0.12)



| Location | 6,975,139 – 6,975,248 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.51 |
| Mean single sequence MFE | -23.88 |
| Consensus MFE | -20.32 |
| Energy contribution | -20.56 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6975139 109 - 23771897 UAGAGUCGUCACACAGUGGCAACAAAAUCG-----------CAACAAAAGGGGAAACUUUCAACAGUCAAGCCCACGAAUGUUAACACAAAUGUGCGGCAAAAAUGUUUGCUGUCACAAC ....((.(((((...))))).)).......-----------.((((...((((..(((......)))....))).)...))))........(((((((((((....))))))).)))).. ( -24.30) >DroSec_CAF1 45046 109 - 1 UAGAGUUGUCACACAGUGGCAACAAAAUCG-----------CAACAAAAGGGGAAACUUUCAACAGUCAAGCCCACGAAUGUUAACACAAAUGUGUGGCAAAAAUGUUUGCUGUCACAAC ..((((((((((...))))))))......(-----------(((((..((((....))))..........(((((((..(((....)))..)))).))).....)).))))..))..... ( -27.00) >DroSim_CAF1 44836 109 - 1 UAGAGUCGUCACACAGUGGCAACAAAAUCG-----------CAACAAAAGGGGAAACUUUCAACAGUCAAGCCCACGAAUGUUAACACAAAUGUGUGGCAAAAAUGUUUGCUGUCACAAC ...............((((((.((((....-----------.......((((....))))..........(((((((..(((....)))..)))).))).......)))).))))))... ( -23.70) >DroEre_CAF1 44981 106 - 1 UAGAG---CCACACAGUGGCAACAAAAUCG-----------CAACAAAAGGGGGAACUUUCAACAGUCAAGCCCACAAAUGUUAACACAAAUGUGUGCCAAAAAUGUUUGCUGUCACAAC .....---.......((((((.((((....-----------.((((.....(((.(((......)))....))).....)))).((((....))))..........)))).))))))... ( -20.50) >DroYak_CAF1 46317 117 - 1 UAGAG---UCACACAGUGGCAACAAAAUCGCAACACAAUCGCAACAAAAGGGGGAACUUUCAACUGUCAAGCCCACGAAUGUUAGCACAAAUGUGUGCCAAAAAUGUUUGCUGUCACAAC .....---.......((((((.((((...(((.((((...((((((...(.(((.((........))....))).)...)))).)).....)))))))........)))).))))))... ( -23.90) >consensus UAGAGU_GUCACACAGUGGCAACAAAAUCG___________CAACAAAAGGGGAAACUUUCAACAGUCAAGCCCACGAAUGUUAACACAAAUGUGUGGCAAAAAUGUUUGCUGUCACAAC ...............((((((.((((......................((((....))))..........(((((((..(((....)))..)))).))).......)))).))))))... (-20.32 = -20.56 + 0.24)


| Location | 6,975,218 – 6,975,327 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.63 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -24.04 |
| Energy contribution | -23.88 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6975218 109 + 23771897 ----------CGAUUUUGUUGCCACUGUGUGACGACUCUAAUUCACGGCAACAGCAACCGGCAGCGACAACUGGCCGAAAAGCAAAUGUACUUUGAUGUAAUACACAUAAACAUU-UGCA ----------.....((((((((.....((((..........))))))))))))....((((((......)).))))....((((((((....((.((.....))))...)))))-))). ( -27.90) >DroSec_CAF1 45125 109 + 1 ----------CGAUUUUGUUGCCACUGUGUGACAACUCUAAUUCACGGCAACAGCAACCGGCAGCGACAACUGGCCGAAAAGCAAAUGUACUUUGAUGUAAUACACAUAAACAUU-UGCA ----------.....((((((((.....((((..........))))))))))))....((((((......)).))))....((((((((....((.((.....))))...)))))-))). ( -27.90) >DroSim_CAF1 44915 109 + 1 ----------CGAUUUUGUUGCCACUGUGUGACGACUCUAAUUCACGGCAACAGCAACCGGCAGCGACAACUGGCCGAAAAGCAAAUGUACUUUGAUGUAAUAAACAUAAACAUU-UGCA ----------.....((((((((.....((((..........))))))))))))....((((((......)).))))....((((((((......((((.....))))..)))))-))). ( -28.10) >DroEre_CAF1 45060 106 + 1 ----------CGAUUUUGUUGCCACUGUGUGG---CUCUAAUUCACGGCAACAGCAACCGGCAGCGACAACUGGCCGAAAAGCAAAUGUACUUUGAUGUAAUACACAUAAACAUU-UGCA ----------.....((((((((.....((((---.......))))))))))))....((((((......)).))))....((((((((....((.((.....))))...)))))-))). ( -27.60) >DroYak_CAF1 46397 117 + 1 GAUUGUGUUGCGAUUUUGUUGCCACUGUGUGA---CUCUAAUUCACGGCAACAGCAACCGGCAGCGACAACUGGCCGAAAAGCAAAUGUACUUUGAUGUAAUACACAUAAACAUUUUGCA ......(((((.....(((((((.....((((---.......))))))))))))))))((((((......)).))))....((((((((....((.((.....))))...))).))))). ( -28.80) >consensus __________CGAUUUUGUUGCCACUGUGUGAC_ACUCUAAUUCACGGCAACAGCAACCGGCAGCGACAACUGGCCGAAAAGCAAAUGUACUUUGAUGUAAUACACAUAAACAUU_UGCA ...............((((((((.....((((..........))))))))))))....((((((......)).))))....((((((((......((((.....))))..))))).))). (-24.04 = -23.88 + -0.16)

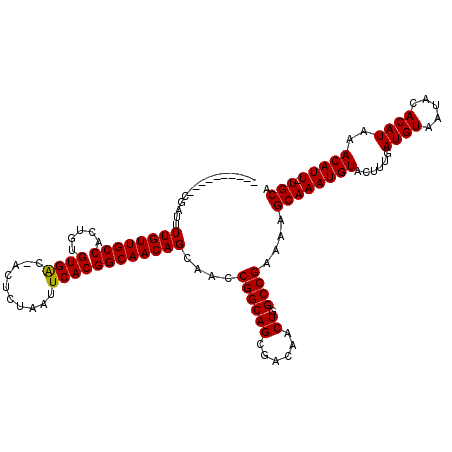
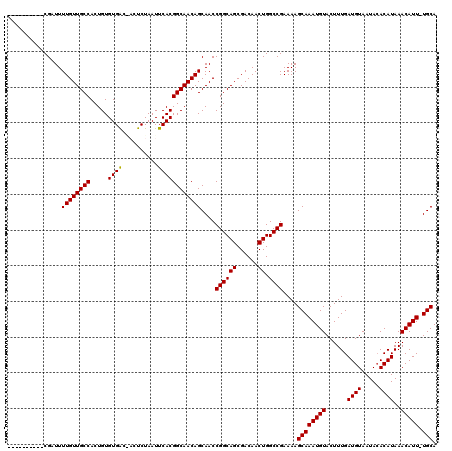
| Location | 6,975,218 – 6,975,327 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.63 |
| Mean single sequence MFE | -31.86 |
| Consensus MFE | -27.22 |
| Energy contribution | -27.42 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.957111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6975218 109 - 23771897 UGCA-AAUGUUUAUGUGUAUUACAUCAAAGUACAUUUGCUUUUCGGCCAGUUGUCGCUGCCGGUUGCUGUUGCCGUGAAUUAGAGUCGUCACACAGUGGCAACAAAAUCG---------- .(((-(((((....(((.....)))......))))))))....((((.(((....)))))))(((((..(((..((((..........)))).)))..))))).......---------- ( -32.30) >DroSec_CAF1 45125 109 - 1 UGCA-AAUGUUUAUGUGUAUUACAUCAAAGUACAUUUGCUUUUCGGCCAGUUGUCGCUGCCGGUUGCUGUUGCCGUGAAUUAGAGUUGUCACACAGUGGCAACAAAAUCG---------- .(((-(((((....(((.....)))......))))))))....((((.(((....)))))))(((((..(((..((((..........)))).)))..))))).......---------- ( -32.30) >DroSim_CAF1 44915 109 - 1 UGCA-AAUGUUUAUGUUUAUUACAUCAAAGUACAUUUGCUUUUCGGCCAGUUGUCGCUGCCGGUUGCUGUUGCCGUGAAUUAGAGUCGUCACACAGUGGCAACAAAAUCG---------- .(((-(((((..((((.....))))......))))))))....((((.(((....)))))))(((((..(((..((((..........)))).)))..))))).......---------- ( -32.30) >DroEre_CAF1 45060 106 - 1 UGCA-AAUGUUUAUGUGUAUUACAUCAAAGUACAUUUGCUUUUCGGCCAGUUGUCGCUGCCGGUUGCUGUUGCCGUGAAUUAGAG---CCACACAGUGGCAACAAAAUCG---------- .(((-(((((....(((.....)))......))))))))....((((.(((....)))))))(((((..(((..(((........---.))).)))..))))).......---------- ( -31.20) >DroYak_CAF1 46397 117 - 1 UGCAAAAUGUUUAUGUGUAUUACAUCAAAGUACAUUUGCUUUUCGGCCAGUUGUCGCUGCCGGUUGCUGUUGCCGUGAAUUAGAG---UCACACAGUGGCAACAAAAUCGCAACACAAUC .............(((((..(((......)))....(((....((((.(((....)))))))(((((..(((..((((.......---)))).)))..)))))......))))))))... ( -31.20) >consensus UGCA_AAUGUUUAUGUGUAUUACAUCAAAGUACAUUUGCUUUUCGGCCAGUUGUCGCUGCCGGUUGCUGUUGCCGUGAAUUAGAGU_GUCACACAGUGGCAACAAAAUCG__________ .(((.(((((..((((.....))))......))))))))....((((.(((....)))))))(((((..(((..((((..........)))).)))..)))))................. (-27.22 = -27.42 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:15 2006