
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,972,090 – 6,972,227 |
| Length | 137 |
| Max. P | 0.928395 |

| Location | 6,972,090 – 6,972,187 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.19 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -17.08 |
| Energy contribution | -17.76 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6972090 97 + 23771897 UGGUCGGUGCAAAGAC---GACAGCAGGGAGAAGUCGGAAAAUGCUGAAGCCAUAAGCACCUGCAGUCGCUGCACCUUGCAGCGGAAAAUUGC--------------------CGCUUUG .....((((((..(.(---(((.(((((......((((......)))).((.....)).))))).)))))))))))....(((((.......)--------------------))))... ( -34.40) >DroSec_CAF1 42039 97 + 1 UGGUUGGUGCAAAGAC---GACAGCAGGGAAAAGUCGGAAAAUGCUGAAGCCAUAAGCACCUGCAGUCGCUGCACCAAGCAGCGGAAAAUUGC--------------------CGCUUUG ((.((((((((..(.(---(((.(((((......((((......)))).((.....)).))))).))))))))))))).))((((.......)--------------------))).... ( -38.70) >DroSim_CAF1 41778 97 + 1 UGGUUGGUGCAAAGAC---GACAGCAGGGAAAAGUCGGAAAAUGCUGAAGCCAUAAGCACCUGCAGUCGCAGCACCAUGCAGCGGAAAAUUGC--------------------CGCUUUG ..((((((((...(.(---(((.(((((......((((......)))).((.....)).))))).))))).)))))).))(((((.......)--------------------))))... ( -34.50) >DroEre_CAF1 42060 91 + 1 UGGUCGCCGCCAAGAC---GACAGCGGGGAAAAGUCGGAAAAUGCAGAAGCCUUAAGCACCUGCAGUCACAACACCAUGCAGCGGAAAAUUGCA-------------------------- (((((.((((......---....)))).))...((.(((...(((((..((.....))..))))).)).).)).)))(((((.......)))))-------------------------- ( -22.80) >DroYak_CAF1 43211 120 + 1 UGGUCGCCGCAAAGACGACGACAGCUGGGAAAAGUCGGAAAAUGCAGAAGCCCUAAGCACCUGCAGUCACAGUCCCAUGCAGUGGAAAAUUGCGCUGCAACUGGCGAAAUUGCGCCUUUG ..((((.((......)).))))...(((((.......((...(((((..((.....))..))))).))....)))))(((((((........)))))))...((((......)))).... ( -39.20) >consensus UGGUCGGUGCAAAGAC___GACAGCAGGGAAAAGUCGGAAAAUGCUGAAGCCAUAAGCACCUGCAGUCGCAGCACCAUGCAGCGGAAAAUUGC____________________CGCUUUG ........((((.......(((.(((((......((((......)))).((.....)).))))).)))((.((.....)).))......))))........................... (-17.08 = -17.76 + 0.68)



| Location | 6,972,090 – 6,972,187 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.19 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -16.42 |
| Energy contribution | -17.18 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6972090 97 - 23771897 CAAAGCG--------------------GCAAUUUUCCGCUGCAAGGUGCAGCGACUGCAGGUGCUUAUGGCUUCAGCAUUUUCCGACUUCUCCCUGCUGUC---GUCUUUGCACCGACCA ....(((--------------------((........)))))..(((((((((((.(((((((((.(.....).))))......((....))))))).)))---))...))))))..... ( -35.00) >DroSec_CAF1 42039 97 - 1 CAAAGCG--------------------GCAAUUUUCCGCUGCUUGGUGCAGCGACUGCAGGUGCUUAUGGCUUCAGCAUUUUCCGACUUUUCCCUGCUGUC---GUCUUUGCACCAACCA ..(((((--------------------((........)))))))(((((((((((.(((((((((.(.....).))))......((....))))))).)))---))...))))))..... ( -36.40) >DroSim_CAF1 41778 97 - 1 CAAAGCG--------------------GCAAUUUUCCGCUGCAUGGUGCUGCGACUGCAGGUGCUUAUGGCUUCAGCAUUUUCCGACUUUUCCCUGCUGUC---GUCUUUGCACCAACCA ....(((--------------------((........))))).((((((.(((((.(((((((((.(.....).))))......((....))))))).)))---))....)))))).... ( -34.60) >DroEre_CAF1 42060 91 - 1 --------------------------UGCAAUUUUCCGCUGCAUGGUGUUGUGACUGCAGGUGCUUAAGGCUUCUGCAUUUUCCGACUUUUCCCCGCUGUC---GUCUUGGCGGCGACCA --------------------------((((.........))))((((((((.((.((((((.((.....)).))))))...)))))).......(((((((---.....))))))))))) ( -29.00) >DroYak_CAF1 43211 120 - 1 CAAAGGCGCAAUUUCGCCAGUUGCAGCGCAAUUUUCCACUGCAUGGGACUGUGACUGCAGGUGCUUAGGGCUUCUGCAUUUUCCGACUUUUCCCAGCUGUCGUCGUCUUUGCGGCGACCA ....((((......))))((((((((.(........).)))))(((((..((((.((((((.((.....)).))))))...))..))...))))))))((((((((....)))))))).. ( -39.40) >consensus CAAAGCG____________________GCAAUUUUCCGCUGCAUGGUGCUGCGACUGCAGGUGCUUAUGGCUUCAGCAUUUUCCGACUUUUCCCUGCUGUC___GUCUUUGCACCGACCA ...........................(((.........)))..(((((((.((((((.((.((.....)).)).)))......(((...........)))...))).)))))))..... (-16.42 = -17.18 + 0.76)

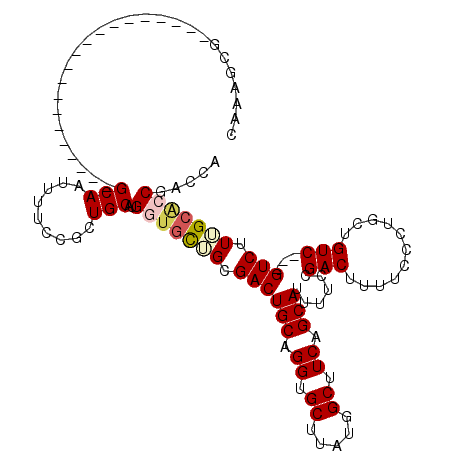

| Location | 6,972,127 – 6,972,227 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.48 |
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -18.47 |
| Energy contribution | -18.71 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6972127 100 + 23771897 AAUGCUGAAGCCAUAAGCACCUGCAGUCGCUGCACCUUGCAGCGGAAAAUUGC--------------------CGCUUUGAUUUGGCUUGCAGGAAAUUCGCGUUACUGGAAAUAUUGUU (((((.(((((.....)).((((((((((((((.....)))))))......((--------------------((........)))))))))))...))))))))............... ( -32.20) >DroSec_CAF1 42076 100 + 1 AAUGCUGAAGCCAUAAGCACCUGCAGUCGCUGCACCAAGCAGCGGAAAAUUGC--------------------CGCUUUGAUUUGACUUGCAGGAAAUUCGCGUUACUGGAAAUAUUGUU (((((.(((((.....)).((((((((((((......)))(((((.......)--------------------)))).......))).))))))...))))))))............... ( -30.50) >DroSim_CAF1 41815 100 + 1 AAUGCUGAAGCCAUAAGCACCUGCAGUCGCAGCACCAUGCAGCGGAAAAUUGC--------------------CGCUUUGAUUUGACCUGCAGGAAAUUCGCGUUACUGGAAAUAUUGUU (((((.(((((.....)).((((((((((((......)))(((((.......)--------------------)))).......)).)))))))...))))))))............... ( -30.20) >DroYak_CAF1 43251 111 + 1 AAUGCAGAAGCCCUAAGCACCUGCAGUCACAGUCCCAUGCAGUGGAAAAUUGCGCUGCAACUGGCGAAAUUGCGCCUUUGAUUUCACUCGCAGGA---------UACUUGAAGUAUUGCU ...((((..((..((((..((((((((...((((...(((((((........)))))))...((((......))))...))))..))).))))).---------..))))..)).)))). ( -32.40) >consensus AAUGCUGAAGCCAUAAGCACCUGCAGUCGCAGCACCAUGCAGCGGAAAAUUGC____________________CGCUUUGAUUUGACUUGCAGGAAAUUCGCGUUACUGGAAAUAUUGUU ..((((.........))))((((((((((((((.....))))))).(((((............................)))))...))))))).......................... (-18.47 = -18.71 + 0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:04 2006