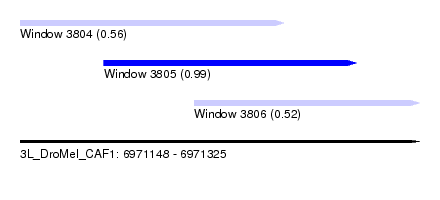
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,971,148 – 6,971,325 |
| Length | 177 |
| Max. P | 0.992664 |

| Location | 6,971,148 – 6,971,265 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.97 |
| Mean single sequence MFE | -40.27 |
| Consensus MFE | -27.47 |
| Energy contribution | -28.11 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.556070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6971148 117 + 23771897 UAGGCAAAACAUUUUUCU-CAGUGUCUGAGACGCAGGC--CGGCUUCGGAGUUGGAAAAAAAAUGGCCAGACAAACACAUCGCCGGCCAAAUGGAUGCACAGGCAUGCCGGCGAAGGCAG ..(((....(((((.(((-(((...))))))....(((--((((...((.(((........)))..)).((........)))))))))))))).((((....))))))).((....)).. ( -38.90) >DroSec_CAF1 40094 106 + 1 -----------UUUUUCU-CAGUGUCUGAGGCGCAGGC--CGGCUUCGGAGCCGGGAAAAAAAUGGCCAGACAAACACAUCGCCGGCCAAAUGGAUGCACAGGCAUGCCGGCGAAGGCAG -----------(((((((-(.((.((((((((......--..)))))))))).))))))))....(((.(.....)...((((((((.......((((....)))))))))))).))).. ( -40.81) >DroSim_CAF1 40855 106 + 1 -----------UUUUUCU-CAGUGUCUGAGGCGGAGGC--CGGCUUCGGAGCCGGGAAAAAAAUGGCCAGACAAACACAUCGCCGGCCAAAUGGAUGCACAGGCAUGCCGGCGAAGGCAG -----------(((((((-(.((.((((((((.(....--).)))))))))).))))))))....(((.(.....)...((((((((.......((((....)))))))))))).))).. ( -42.11) >DroEre_CAF1 41136 118 + 1 CAGGUUGAUCAUUUCUCU-CAGUGUCUUUGGCGGAGGCUCUGGCUCUGGAGCCGGGAA-AAAAUAUCCAGACAAACACAUCGCCGGCCAAAUGGAUGCACAGGCAUGCCGGCGAAGGCAG ..((..((.....))..)-)..((((((((.(((..((.(((((((((..(((((((.-....................)).)))))....)))).)).)))))...))).)))))))). ( -37.60) >DroYak_CAF1 42235 118 + 1 CAAGCAGAUAUUUUUUUUCCAGUGUCUUGGGCGGAGGC--UGGCUUUGGAGGCGGAAAAAAAAUGUCCAGACAAACACAUCGGCGGCCAAAUGGAUGCACAGGCAUGCCGGCGAAGGCAG ...((.((((((((((((((.((.(((..(((......--..)))..))).))))))))))))))))..........(.(((.((((.......((((....)))))))).))).))).. ( -41.91) >consensus _A_G___A___UUUUUCU_CAGUGUCUGAGGCGGAGGC__CGGCUUCGGAGCCGGGAAAAAAAUGGCCAGACAAACACAUCGCCGGCCAAAUGGAUGCACAGGCAUGCCGGCGAAGGCAG ......................((((((...(((((.(...).)))))..((((.........))))))))))....(.((((((((.......((((....)))))))))))).).... (-27.47 = -28.11 + 0.64)

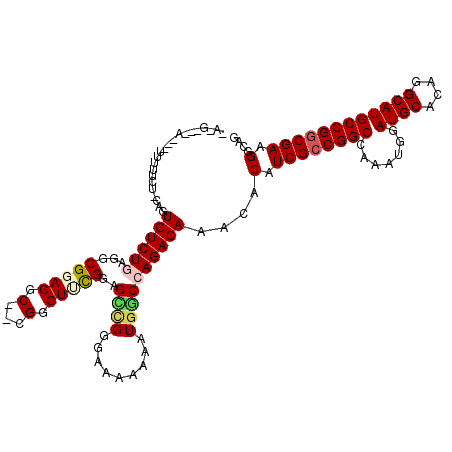
| Location | 6,971,185 – 6,971,297 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.22 |
| Mean single sequence MFE | -39.99 |
| Consensus MFE | -34.79 |
| Energy contribution | -34.87 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6971185 112 + 23771897 CGGCUUCGGAGUUGGAAAAAAAAUGGCCAGACAAACACAUCGCCGGCCAAAUGGAUGCACAGGCAUGCCGGCGAAGGCAGUUCAAGUGCACAUGGCCAACAUGAGCUGCGAU-------- .((((......((....)).....)))).........(.((((((((.......((((....)))))))))))).)((((((((.((((.....))..)).))))))))...-------- ( -38.81) >DroSec_CAF1 40120 112 + 1 CGGCUUCGGAGCCGGGAAAAAAAUGGCCAGACAAACACAUCGCCGGCCAAAUGGAUGCACAGGCAUGCCGGCGAAGGCAGUUCAAGUGCACAUGGCCAACAUGAGCUGCGAU-------- (((((....))))).......................(.((((((((.......((((....)))))))))))).)((((((((.((((.....))..)).))))))))...-------- ( -39.91) >DroSim_CAF1 40881 112 + 1 CGGCUUCGGAGCCGGGAAAAAAAUGGCCAGACAAACACAUCGCCGGCCAAAUGGAUGCACAGGCAUGCCGGCGAAGGCAGUUCAAGUGCACAUGGCCAACAUGAGCUGCGAG-------- (((((....))))).......................(.((((((((.......((((....)))))))))))).)((((((((.((((.....))..)).))))))))...-------- ( -39.91) >DroEre_CAF1 41175 119 + 1 UGGCUCUGGAGCCGGGAA-AAAAUAUCCAGACAAACACAUCGCCGGCCAAAUGGAUGCACAGGCAUGCCGGCGAAGGCAGUUCAAGUGCACAUGGCCAACAUGAGCUGCGAUUCUGCGAU .((((....)))).....-.....(((((((......(.((((((((.......((((....)))))))))))).)((((((((.((((.....))..)).))))))))...)))).))) ( -43.51) >DroYak_CAF1 42273 120 + 1 UGGCUUUGGAGGCGGAAAAAAAAUGUCCAGACAAACACAUCGGCGGCCAAAUGGAUGCACAGGCAUGCCGGCGAAGGCAGUUCAAGUGCACAUAGGCAACAUGAGCUGCAAUUCUGCGAU ...........((((((......(((....)))....(.(((.((((.......((((....)))))))).))).)((((((((..(((......)))...))))))))..))))))... ( -37.81) >consensus CGGCUUCGGAGCCGGGAAAAAAAUGGCCAGACAAACACAUCGCCGGCCAAAUGGAUGCACAGGCAUGCCGGCGAAGGCAGUUCAAGUGCACAUGGCCAACAUGAGCUGCGAU________ (((((....))))).......................(.((((((((.......((((....)))))))))))).)((((((((.((((.....))..)).))))))))........... (-34.79 = -34.87 + 0.08)

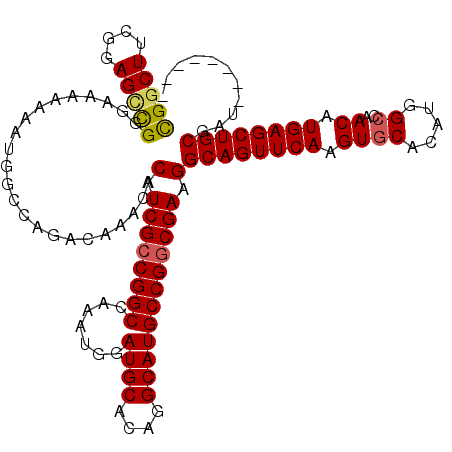
| Location | 6,971,225 – 6,971,325 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.58 |
| Mean single sequence MFE | -37.41 |
| Consensus MFE | -28.01 |
| Energy contribution | -28.41 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6971225 100 + 23771897 CGCCGGCCAAAUGGAUGCACAGGCAUGCCGGCGAAGGCAGUUCAAGUGCACAUGGCCAACAUGAGCUGCGAU--------GCU--------GCGAUGCUGU----GAUGCUGCUUGAAUA (((((((.......((((....))))))))))).((((((((((...(((((((.....))))(((......--------)))--------....)))..)----)).)))))))..... ( -37.61) >DroSec_CAF1 40160 92 + 1 CGCCGGCCAAAUGGAUGCACAGGCAUGCCGGCGAAGGCAGUUCAAGUGCACAUGGCCAACAUGAGCUGCGAU------------------------GCUGU----GAUGCUGCUUGAAUA (((((((.......((((....)))))))))))......(((((((((((((((.....))))(((......------------------------)))..----..))).)))))))). ( -34.61) >DroSim_CAF1 40921 96 + 1 CGCCGGCCAAAUGGAUGCACAGGCAUGCCGGCGAAGGCAGUUCAAGUGCACAUGGCCAACAUGAGCUGCGAG------------------------CUUCUAGAGAAUGCUGCUUGAAUA (((((((.......((((....)))))))))))......((((((((((.((((.....)))).))....((------------------------((((....))).))))))))))). ( -35.31) >DroEre_CAF1 41214 108 + 1 CGCCGGCCAAAUGGAUGCACAGGCAUGCCGGCGAAGGCAGUUCAAGUGCACAUGGCCAACAUGAGCUGCGAUUCUGCGAUGCU--------GCGAUGCUGU----GAUGCUGCUUGAAUA (((((((.......((((....))))))))))).((((((((((...(((((((.....))))(((((((....))))..)))--------....)))..)----)).)))))))..... ( -38.81) >DroYak_CAF1 42313 116 + 1 CGGCGGCCAAAUGGAUGCACAGGCAUGCCGGCGAAGGCAGUUCAAGUGCACAUAGGCAACAUGAGCUGCAAUUCUGCGAUGCUGCGAGGCUGCGAUGCUGU----GAUGCUGCUUGAAUA ((((((((...((......))))).)))))((....)).((((((((((((((((.((.(...(((((((....))))..)))(((....)))).))))))----).))).)))))))). ( -40.70) >consensus CGCCGGCCAAAUGGAUGCACAGGCAUGCCGGCGAAGGCAGUUCAAGUGCACAUGGCCAACAUGAGCUGCGAU________GCU________GCGAUGCUGU____GAUGCUGCUUGAAUA (((((((.......((((....)))))))))))...((((((((.((((.....))..)).))))))))................................................... (-28.01 = -28.41 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:02 2006