
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
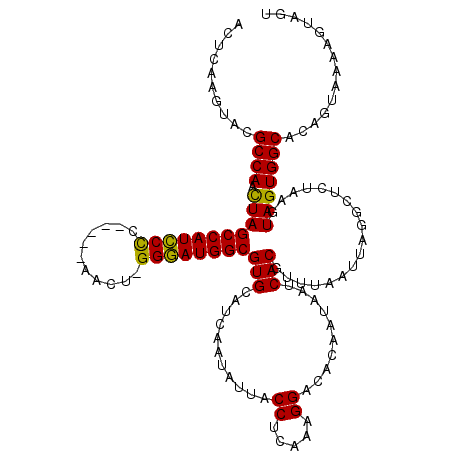
| Location | 6,968,203 – 6,968,318 |
| Length | 115 |
| Max. P | 0.718888 |

| Location | 6,968,203 – 6,968,318 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.64 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -24.78 |
| Energy contribution | -24.14 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
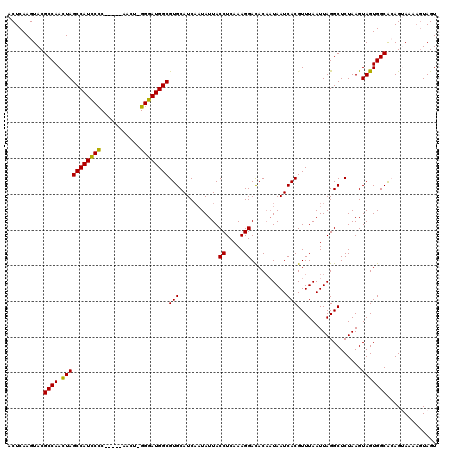
>3L_DroMel_CAF1 6968203 115 + 23771897 ACUCAAGUACGCCAACUAGCCAUCCUC-----AACUUGGGAUGGCGUGCAUCAAUAUUACCUCAAAGGACACAAUAAUCACGUUUAAUUAGGCUCUAAGUAGUGGCACAGUAAAAGUAGU .......(((....(((.((((((((.-----.....))))))))((((..........((.....))..........(((......((((...))))...))))))))))....))).. ( -26.20) >DroSec_CAF1 37613 114 + 1 ACUCAAGUACGCCAACUAGCCAUCCCC-----CACU-GGGAUGGCGUGCAUCAAUAUUACCUCAAAGGACACAAUAAUCACGUUUAAUUAGGCUCUAAGUAGUGGCACAGUAAAUGUAGU (((((..(((........((((((((.-----....-))))))))((((..........((.....))..........(((......((((...))))...))))))).)))..)).))) ( -29.50) >DroSim_CAF1 38454 114 + 1 ACUCAAGUACGCCAACUAGCCAUCCCC-----AACU-GGGAUGGCGUGCAUCAAUAUUACCUCAAAGGAUACAAUAAUCACGUUUAAUUAGGCUCUAAGUAGUGGCACAGUAAAAAUAGU ..............((((((((((((.-----....-))))))))((((..........((.....))..........(((......((((...))))...)))))))........)))) ( -27.10) >DroEre_CAF1 38682 114 + 1 CCUCAAGUACGCCAACUAGCCAUCCCC-----AACU-GGGAUGGCGUGCAUCUUUAUUACCUCAAAGGACACCAUAAUCACGUUUAAUUAGGCUCUAAGUAGUGGCAGAGAACCAGUAGU ..........((((.(((((((((((.-----....-))))))))(((..(((((........))))).)))...........................))))))).............. ( -27.10) >DroYak_CAF1 39752 119 + 1 CCUCAAGUACGCCAAUUAGCCAUUCCCCCGAAAAUU-GGAAUGGCGUGCAUCAAUAUUACCUCAAAGGACACAUUAAUCACGUUUAAUUAGGCUCUAAGUAGUGGCGGAGAAGAAGUAGU .......(((((((((((((((((((..........-))))))))(((.((.(((....((.....))....))).)))))...))))).)))(((..((....))..)))....))).. ( -25.70) >consensus ACUCAAGUACGCCAACUAGCCAUCCCC_____AACU_GGGAUGGCGUGCAUCAAUAUUACCUCAAAGGACACAAUAAUCACGUUUAAUUAGGCUCUAAGUAGUGGCACAGUAAAAGUAGU ..........((((.(((((((((((...........))))))))(((...........((.....))..........)))..................))))))).............. (-24.78 = -24.14 + -0.64)

| Location | 6,968,203 – 6,968,318 |
|---|---|
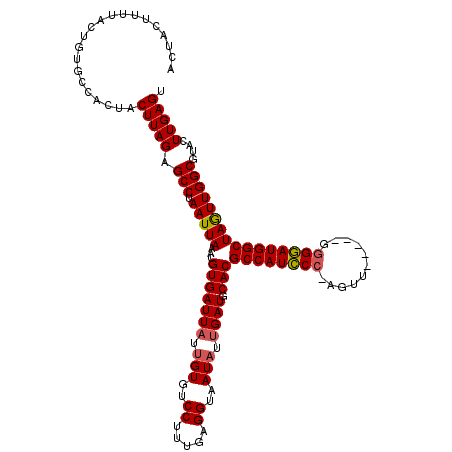
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.64 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -27.38 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
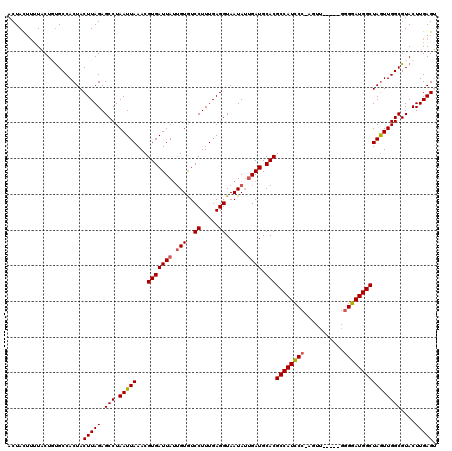
>3L_DroMel_CAF1 6968203 115 - 23771897 ACUACUUUUACUGUGCCACUACUUAGAGCCUAAUUAAACGUGAUUAUUGUGUCCUUUGAGGUAAUAUUGAUGCACGCCAUCCCAAGUU-----GAGGAUGGCUAGUUGGCGUACUUGAGU ...((((.....((((..((....)).(((.(((((...(((((((.(((..((.....))..))).)))).)))((((((((.....-----).)))))))))))))))))))..)))) ( -29.80) >DroSec_CAF1 37613 114 - 1 ACUACAUUUACUGUGCCACUACUUAGAGCCUAAUUAAACGUGAUUAUUGUGUCCUUUGAGGUAAUAUUGAUGCACGCCAUCCC-AGUG-----GGGGAUGGCUAGUUGGCGUACUUGAGU .....(((((..((((..((....)).(((.(((((...(((((((.(((..((.....))..))).)))).)))((((((((-....-----.)))))))))))))))))))).))))) ( -32.90) >DroSim_CAF1 38454 114 - 1 ACUAUUUUUACUGUGCCACUACUUAGAGCCUAAUUAAACGUGAUUAUUGUAUCCUUUGAGGUAAUAUUGAUGCACGCCAUCCC-AGUU-----GGGGAUGGCUAGUUGGCGUACUUGAGU ....................((((((.(((.(((((...(((((((.(((..((.....))..))).)))).)))((((((((-....-----.))))))))))))))))....)))))) ( -32.00) >DroEre_CAF1 38682 114 - 1 ACUACUGGUUCUCUGCCACUACUUAGAGCCUAAUUAAACGUGAUUAUGGUGUCCUUUGAGGUAAUAAAGAUGCACGCCAUCCC-AGUU-----GGGGAUGGCUAGUUGGCGUACUUGAGG .((...(((....(((((((((((((((((((((((....)))))).))....)))))))...............((((((((-....-----.))))))))))).))))).)))..)). ( -34.50) >DroYak_CAF1 39752 119 - 1 ACUACUUCUUCUCCGCCACUACUUAGAGCCUAAUUAAACGUGAUUAAUGUGUCCUUUGAGGUAAUAUUGAUGCACGCCAUUCC-AAUUUUCGGGGGAAUGGCUAAUUGGCGUACUUGAGG ....((((.....(((((.....................(((((((((((..((.....))..)))))))).)))((((((((-..........))))))))....))))).....)))) ( -33.70) >consensus ACUACUUUUACUGUGCCACUACUUAGAGCCUAAUUAAACGUGAUUAUUGUGUCCUUUGAGGUAAUAUUGAUGCACGCCAUCCC_AGUU_____GGGGAUGGCUAGUUGGCGUACUUGAGU .....................(((((.(((.(((((...(((((((.(((..((.....))..))).)))).)))((((((((...........))))))))))))))))....))))). (-27.38 = -27.50 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:58 2006