
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
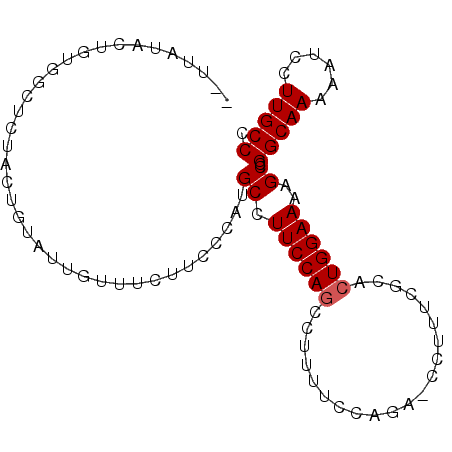
| Location | 6,953,187 – 6,953,280 |
| Length | 93 |
| Max. P | 0.999567 |

| Location | 6,953,187 – 6,953,280 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 76.53 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -10.80 |
| Energy contribution | -11.00 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6953187 93 + 23771897 --UUAUACUGUAACUCUACUGUAUUGUUUCAUCCCAUGCCUUCCAGCCUUUUCCAGACCCUUUCGCACUGGAAAAGCCGGCAAAAAUCCUUGCCC --..((((.(((....))).)))).....................(.(((((((((...........))))))))).)(((((......))))). ( -19.00) >DroSec_CAF1 22793 93 + 1 --UUGUAAUGUGGCUCUAUUGUAUUGUUUCUUCCCAUGCCUUCCAGCCUUUUCCAGAUCCUUUCGCACUGGAAAAGCCGGCAAAAAUCCUUGCCC --.........(((.......................))).....(.(((((((((...........))))))))).)(((((......))))). ( -19.10) >DroSim_CAF1 23446 93 + 1 --UUAUACUGUGGCUCUACUGUAUUGUUUCUUGCCAUGCCUUCCAGCCUUUUCCAGACCCUUUCGCACUGGAAAAGCCGGCAAAAAUCCUUGCCC --.......(((((...((......)).....)))))........(.(((((((((...........))))))))).)(((((......))))). ( -23.40) >DroEre_CAF1 23527 78 + 1 -----------GGCUCUGCUGUAUUAUUUGUUUCCAGGCCUUCCAGCAUU---GGGA--UUUC-CCACUGGAAAAGCCGGCAAUAAUCCUUGCCC -----------((((...............(((((((((......))..(---(((.--...)-))))))))))))))(((((......))))). ( -24.86) >DroYak_CAF1 23454 89 + 1 CAAUAUACUGCGGUUCUGCUUUAUAAUUUGUUUCGCGGCCUUCCAUUAUG---GGGA--UUUCCCCACUGGAAAAGCCGGCAA-AAUCCUUGCCA .........((((..(.............)..))))(((.(((((...((---(((.--...))))).)))))..)))(((((-.....))))). ( -29.52) >consensus __UUAUACUGUGGCUCUACUGUAUUGUUUCUUCCCAUGCCUUCCAGCCUUUUCCAGA_CCUUUCGCACUGGAAAAGCCGGCAAAAAUCCUUGCCC .....................................((.((((((.....................))))))..)).(((((......))))). (-10.80 = -11.00 + 0.20)

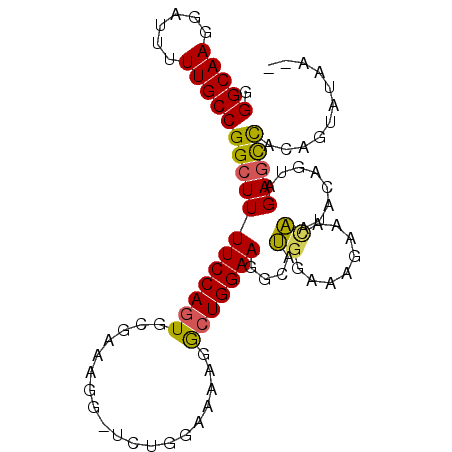
| Location | 6,953,187 – 6,953,280 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 76.53 |
| Mean single sequence MFE | -29.16 |
| Consensus MFE | -15.41 |
| Energy contribution | -15.09 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.53 |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6953187 93 - 23771897 GGGCAAGGAUUUUUGCCGGCUUUUCCAGUGCGAAAGGGUCUGGAAAAGGCUGGAAGGCAUGGGAUGAAACAAUACAGUAGAGUUACAGUAUAA-- .((((((....))))))..((((((((((.(....).).)))))))))(((....))).............((((.(((....))).))))..-- ( -26.00) >DroSec_CAF1 22793 93 - 1 GGGCAAGGAUUUUUGCCGGCUUUUCCAGUGCGAAAGGAUCUGGAAAAGGCUGGAAGGCAUGGGAAGAAACAAUACAAUAGAGCCACAUUACAA-- .(((.....(((((.(((.((((((((((.(....).).)))))))))(((....))).))).)))))......(....).))).........-- ( -26.80) >DroSim_CAF1 23446 93 - 1 GGGCAAGGAUUUUUGCCGGCUUUUCCAGUGCGAAAGGGUCUGGAAAAGGCUGGAAGGCAUGGCAAGAAACAAUACAGUAGAGCCACAGUAUAA-- .(((.....(((((((((.((((((((((.(....).).)))))))))(((....))).)))))))))((......))...))).........-- ( -32.90) >DroEre_CAF1 23527 78 - 1 GGGCAAGGAUUAUUGCCGGCUUUUCCAGUGG-GAAA--UCCC---AAUGCUGGAAGGCCUGGAAACAAAUAAUACAGCAGAGCC----------- .(((((......)))))((((((.(((((((-(...--.)))---...)))))))))))((....)).................----------- ( -29.20) >DroYak_CAF1 23454 89 - 1 UGGCAAGGAUU-UUGCCGGCUUUUCCAGUGGGGAAA--UCCC---CAUAAUGGAAGGCCGCGAAACAAAUUAUAAAGCAGAACCGCAGUAUAUUG .((((((...)-)))))((((((.(((((((((...--.)))---)))..)))))))))...........((((..((......))..))))... ( -30.90) >consensus GGGCAAGGAUUUUUGCCGGCUUUUCCAGUGCGAAAGG_UCUGGAAAAGGCUGGAAGGCAUGGAAAGAAACAAUACAGUAGAGCCACAGUAUAA__ .(((((......)))))((((((((((((...................)))))))....((........))........)))))........... (-15.41 = -15.09 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:46 2006