| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
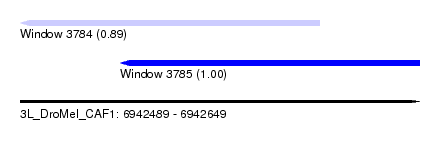
| Location | 6,942,489 – 6,942,649 |
| Length | 160 |
| Max. P | 0.999264 |

| Location | 6,942,489 – 6,942,609 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -40.06 |
| Consensus MFE | -34.13 |
| Energy contribution | -34.45 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6942489 120 - 23771897 CCUAAAAGCCCUCAGGGAACUGGACAGCAAAGUGUUCCACGUUCCACCGCAUGUGCUGCAUCCGGAGCACAUGUUCGUCGAGAAUCAGUAUACCAGCGAAGAGGAGGAGCAGAAGACAGC .......((((((..(((((((((((......)).)))).)))))..((((((((((.(....).)))))))((((.....))))..........))).....)))).)).......... ( -37.60) >DroSec_CAF1 11943 120 - 1 CCUGAAAGCCCUCAGGCAACUGGACAGCAAAGUAUUCCACGUUCCACCGCAUGUGCUGCAUCCGGAGCACAUGUUCUUCGAGAAUCAGUAUACCAGCGAGGAGGAGGAGCAGAAAACAGC .(((...((..((((....))))...))............(((((.(((((((((((.(....).)))))))))((((((................)))))))).)))))......))). ( -39.29) >DroSim_CAF1 12634 120 - 1 CCUAAAAGCCCUCAGGCAACUGGACAGCAAAGUGUUCCACGUUCCACCGCAUGUGCUGCAUCCGGAGCACAUGUUCUUCGAGAAUCAGUAUACCAGCGAGGAGGAGGAGCAGAAGACAGC .......((..((((....))))...))...((.(((...(((((.(((((((((((.(....).)))))))))((((((................)))))))).))))).))).))... ( -40.49) >DroEre_CAF1 12446 120 - 1 CCUAAAAGCCCUGAGGGAACUGGACAGGAAGGUGUUCCACGUUCCAUCGCAUGUGCUGCAUCCGGAGCACAUGUUCUUCGCAAAUCAGUUUACCAGCGAGGAGGAGGAGCAGAAAGUAGC .......(((((((.(((((((((((......)).)))).))))).))(((((((((.(....).)))))))))(((((((..............)))))))..))).)).......... ( -41.84) >DroYak_CAF1 12321 120 - 1 CCUAAAAGCUCUGAGGGAACUGGACAGCCGGGUGUUCCACGUUCCACCGCAUGUGCUGCAUCCGGAGCACAUGUUCUUCGAAAAUCAGUACACCAGCGAGGAGGAGGAGCAGAAGACAGC ..........(((.((((.((((....))))...))))..(((((.(((((((((((.(....).)))))))))((((((................)))))))).)))))......))). ( -41.09) >consensus CCUAAAAGCCCUCAGGGAACUGGACAGCAAAGUGUUCCACGUUCCACCGCAUGUGCUGCAUCCGGAGCACAUGUUCUUCGAGAAUCAGUAUACCAGCGAGGAGGAGGAGCAGAAGACAGC .......((..((((....))))...))...((.(((...(((((.(((((((((((.(....).)))))))))((((((................)))))))).))))).))).))... (-34.13 = -34.45 + 0.32)


| Location | 6,942,529 – 6,942,649 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -49.60 |
| Consensus MFE | -42.92 |
| Energy contribution | -44.00 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.999264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6942529 120 - 23771897 AGACCGUUGAGCGUCGUUUCCAGGCAUCCGCCUCCAAGGGCCUAAAAGCCCUCAGGGAACUGGACAGCAAAGUGUUCCACGUUCCACCGCAUGUGCUGCAUCCGGAGCACAUGUUCGUCG .(((.((.((((((.((((((((((....))))...(((((......)))))..)))))).(((((......))))).)))))).)).(((((((((.(....).)))))))))..))). ( -47.90) >DroSec_CAF1 11983 120 - 1 AGAACGUGGAGCGUCGUUUCCAGGCAUCCGCCUCCAAGGGCCUGAAAGCCCUCAGGCAACUGGACAGCAAAGUAUUCCACGUUCCACCGCAUGUGCUGCAUCCGGAGCACAUGUUCUUCG .(((((((((((...((((((((......((((...(((((......))))).))))..))))).)))...))..)))))))))....(((((((((.(....).)))))))))...... ( -50.40) >DroSim_CAF1 12674 120 - 1 AGACCGUGGAGCGUCGUUUCGAGGCAUCCGCCUCCAAGGGCCUAAAAGCCCUCAGGCAACUGGACAGCAAAGUGUUCCACGUUCCACCGCAUGUGCUGCAUCCGGAGCACAUGUUCUUCG .((.(((((((((((.....(((((....)))))..(((((......)))))(((....)))))).((...)))))))))).))....(((((((((.(....).)))))))))...... ( -50.60) >DroEre_CAF1 12486 120 - 1 AAAACGUGGAGCGUCGGUUCCAGGCCGCCGCCGGCAAAGGCCUAAAAGCCCUGAGGGAACUGGACAGGAAGGUGUUCCACGUUCCAUCGCAUGUGCUGCAUCCGGAGCACAUGUUCUUCG .....((((((((((((((((..((((....))))...(((......))).....)))))))....((((....))))))))))))).(((((((((.(....).)))))))))...... ( -49.60) >DroYak_CAF1 12361 120 - 1 AAACCGUGGAGCGUCGUUUCCAGGCCGCCGCCUGCAAGGGCCUAAAAGCUCUGAGGGAACUGGACAGCCGGGUGUUCCACGUUCCACCGCAUGUGCUGCAUCCGGAGCACAUGUUCUUCG .....(((((((((......(((((....)))))..(((((......)))))..((((.((((....))))...))))))))))))).(((((((((.(....).)))))))))...... ( -49.50) >consensus AGACCGUGGAGCGUCGUUUCCAGGCAUCCGCCUCCAAGGGCCUAAAAGCCCUCAGGGAACUGGACAGCAAAGUGUUCCACGUUCCACCGCAUGUGCUGCAUCCGGAGCACAUGUUCUUCG .....(((((((((.((((((.(((....)))....(((((......)))))..)))))).(((((......))))).))))))))).(((((((((.(....).)))))))))...... (-42.92 = -44.00 + 1.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:42 2006