
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,935,558 – 6,935,672 |
| Length | 114 |
| Max. P | 0.965815 |

| Location | 6,935,558 – 6,935,672 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 84.19 |
| Mean single sequence MFE | -20.87 |
| Consensus MFE | -17.90 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6935558 114 + 23771897 AUUUGUGGUUAUUUUAUGGCUAUUUAACUAACAAAUCUAUGCUUUAAUAGGCAGUUAUUUUUAAGCUAUGACAUGAUAUUAGCCAUAAUUCAAGCUUGUGAAGCUUUAUGAUCA .....(((((((.(((((((((.................(((((....)))))(((((.........))))).......)))))))))...(((((.....))))).))))))) ( -23.80) >DroSec_CAF1 5155 97 + 1 ----------AUUUUAUGGCUAUUUAACUA----UGCUAUGCUUUAAUAGGCAGUUAUUUUUAAGCUAUGACAUGAUAUUAGCCAUAAUUCAAGCUUAUGAAGCUUUAGGA--- ----------...(((((((((......((----((.(((((((.(((((....)))))...)))).))).))))....)))))))))...(((((.....))))).....--- ( -20.50) >DroSim_CAF1 5793 96 + 1 ----------AUUUUAUGGCUAUUUAACUAACAAAUCUAUGCUUUAAUAGGCAGUUAUUUUUAAGCUAU-----GAUAUUAGCCAUAAUUCAAGCUUAUGAAGCUUUAUGA--- ----------...(((((((((.................(((((....)))))(((((.........))-----)))..)))))))))...(((((.....))))).....--- ( -18.30) >consensus __________AUUUUAUGGCUAUUUAACUAACAAAUCUAUGCUUUAAUAGGCAGUUAUUUUUAAGCUAUGACAUGAUAUUAGCCAUAAUUCAAGCUUAUGAAGCUUUAUGA___ .............(((((((((..................((((....))))((((.......))))............)))))))))...(((((.....)))))........ (-17.90 = -17.90 + 0.00)

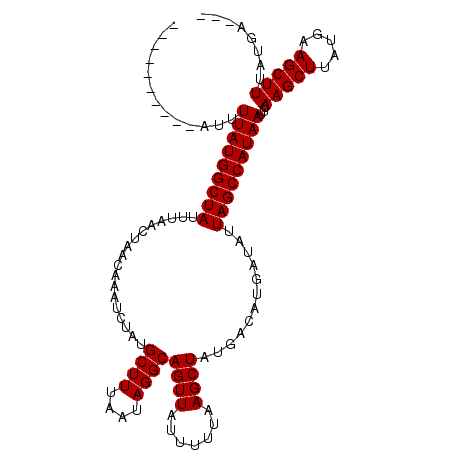

| Location | 6,935,558 – 6,935,672 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 84.19 |
| Mean single sequence MFE | -19.87 |
| Consensus MFE | -16.07 |
| Energy contribution | -16.07 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6935558 114 - 23771897 UGAUCAUAAAGCUUCACAAGCUUGAAUUAUGGCUAAUAUCAUGUCAUAGCUUAAAAAUAACUGCCUAUUAAAGCAUAGAUUUGUUAGUUAAAUAGCCAUAAAAUAACCACAAAU ........(((((.....)))))...(((((((((...........(((((...((((...(((........)))...))))...)))))..)))))))))............. ( -19.02) >DroSec_CAF1 5155 97 - 1 ---UCCUAAAGCUUCAUAAGCUUGAAUUAUGGCUAAUAUCAUGUCAUAGCUUAAAAAUAACUGCCUAUUAAAGCAUAGCA----UAGUUAAAUAGCCAUAAAAU---------- ---.....(((((.....)))))...(((((((((..((.(((..((.((((...((((......)))).))))))..))----).))....)))))))))...---------- ( -20.10) >DroSim_CAF1 5793 96 - 1 ---UCAUAAAGCUUCAUAAGCUUGAAUUAUGGCUAAUAUC-----AUAGCUUAAAAAUAACUGCCUAUUAAAGCAUAGAUUUGUUAGUUAAAUAGCCAUAAAAU---------- ---.....(((((.....)))))...(((((((((.....-----.(((((...((((...(((........)))...))))...)))))..)))))))))...---------- ( -20.50) >consensus ___UCAUAAAGCUUCAUAAGCUUGAAUUAUGGCUAAUAUCAUGUCAUAGCUUAAAAAUAACUGCCUAUUAAAGCAUAGAUUUGUUAGUUAAAUAGCCAUAAAAU__________ ........(((((.....)))))...(((((((((...........(((((..........(((........)))..........)))))..)))))))))............. (-16.07 = -16.07 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:40 2006