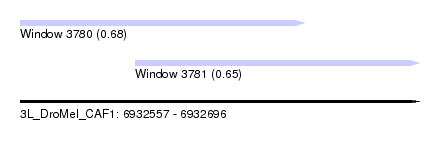
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,932,557 – 6,932,696 |
| Length | 139 |
| Max. P | 0.683786 |

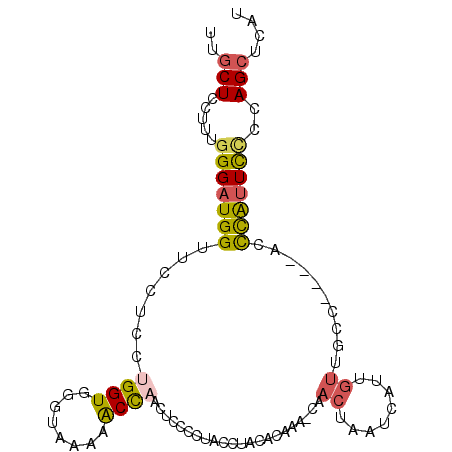
| Location | 6,932,557 – 6,932,656 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 69.49 |
| Mean single sequence MFE | -20.29 |
| Consensus MFE | -6.77 |
| Energy contribution | -6.80 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6932557 99 + 23771897 UAGCUCCUUUGGGAUGGUUCCUCCUGGUGGGUAGACACCAACUCCUCUACCUAUACACA-CAACUAAUCCUUGUUGUCCAUUAACCAAUCCCCAGCUCAU .((((.....(((((((((.......(((((((((..........)))))))))....(-((((........))))).....)))).))))).))))... ( -26.50) >DroSec_CAF1 2146 99 + 1 UUGCUUCUUUGGGAUGGUUCCUCCUGGUGGGUAAACACCAACUCCUCUACCUACACAAA-CAACUAAUCCUUGUUGCUAAUGAUCCAUUCUCCAGCUCAU ..(((.....(((((((.((....(((((((((..............))))))).))..-((((........)))).....)).)))))))..))).... ( -20.84) >DroSim_CAF1 2150 99 + 1 UUGCUCCUUUGGGAUGGUUCCUCCUGGUGGGUAAAUGCCAACUCCACUACCUACACAAA-CAACUAAUCCUUGUUGCUAAUAAUCCAUUCCCCAGCUCAU ..(((.....(((((((.......(((((((((..............))))))).))..-((((........))))........)))))))..))).... ( -21.24) >DroEre_CAF1 2386 95 + 1 UUGCUCCACUGGGAUGGUUCCUCCUGGUGUGUCAAAACUCGCCUCCUUAGCUCCAUAAG-CCACUAACCAUUGUUGCC----ACCCAUUUCCUAGCUUAU ..(((....((((.((((..(....((((.((....)).))))......(((.....))-)...........)..)))----)))))......))).... ( -20.00) >DroYak_CAF1 2385 96 + 1 UUGCUCCAUUGGGAUGGUUCCUCCUGGUGGGUCAAAACUUGCGCCCGUAGCUGCAUAAUAUUACUAAUCAUUGUUGCC----ACCCAUUCCCUAGCUUAU ..(((....((((.((((..(...((((.(((.......(((((.....)).))).......))).))))..)..)))----)))))......))).... ( -22.14) >DroAna_CAF1 2171 87 + 1 UCACUGGUCUUGGAUGGUUCCUGCUGGUAUCCUAAAACUAAAUCCCUUUCUUAUUCUCU-GGUCUAACUAUU------------UUGUUUCCUAGCUUAU ..((..((...(((....))).))..)).............................((-((...(((....------------..)))..))))..... ( -11.00) >consensus UUGCUCCUUUGGGAUGGUUCCUCCUGGUGGGUAAAAACCAACUCCCCUACCUACACAAA_CAACUAAUCAUUGUUGCC____ACCCAUUCCCCAGCUCAU ..(((.....(((((((.......((((........))))......................((........))..........)))))))..))).... ( -6.77 = -6.80 + 0.03)

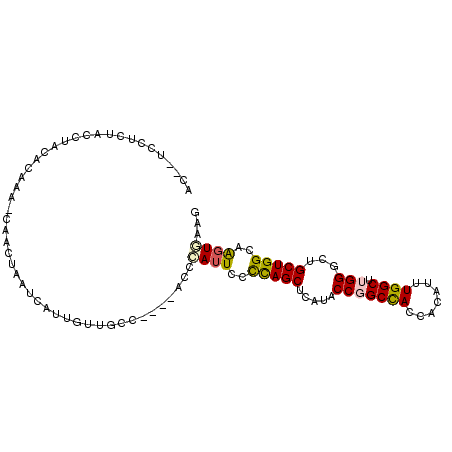
| Location | 6,932,597 – 6,932,696 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 74.13 |
| Mean single sequence MFE | -19.77 |
| Consensus MFE | -14.13 |
| Energy contribution | -14.05 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6932597 99 + 23771897 AC--UCCUCUACCUAUACACA-CAACUAAUCCUUGUUGUCCAUUAACCAAUCCCCAGCUCAUACCGGCCACCACUUUUGGCUUGGGCUGCUGGCAAGUGAAG ..--.............((((-((((........)))))..............(((((.....(((((((.......)))).)))...)))))...)))... ( -23.10) >DroSec_CAF1 2186 99 + 1 AC--UCCUCUACCUACACAAA-CAACUAAUCCUUGUUGCUAAUGAUCCAUUCUCCAGCUCAUACCGGCCACCACUUUUGGCUUGGGCUGCUGGCAAGUUAAG ..--.................-.............((((((..((.....))..(((((((....(((((.......)))))))))))).))))))...... ( -19.50) >DroSim_CAF1 2190 99 + 1 AC--UCCACUACCUACACAAA-CAACUAAUCCUUGUUGCUAAUAAUCCAUUCCCCAGCUCAUACCGGCCACCACUUUUGGCUUGGGCUGCUGGCAAGUAAAG ..--.........(((.....-((((........))))...............(((((.....(((((((.......)))).)))...)))))...)))... ( -19.90) >DroEre_CAF1 2426 95 + 1 GC--CUCCUUAGCUCCAUAAG-CCACUAACCAUUGUUGCC----ACCCAUUUCCUAGCUUAUUCCCGCCACCACAUUUGGCUUGGGCUGCUGGCAAGUGAAG ..--................(-(.((........)).)).----...(((((.(((((.....(((((((.......))))..)))..))))).)))))... ( -19.70) >DroYak_CAF1 2425 96 + 1 GC--GCCCGUAGCUGCAUAAUAUUACUAAUCAUUGUUGCC----ACCCAUUCCCUAGCUUAUUCCGGCCACCACAUUUGGCUUGGGCUGCUGGCAGGUGAAG ..--(((.(((((.(((((((..........)))).))).----...................(((((((.......)))).)))))))).)))........ ( -22.10) >DroPer_CAF1 29182 89 + 1 GCAGUCUUCAAAUUAUA--UU-GUUUUAACCAAU----------UCCUAUUUCUUAGCUCAUACCCGCUACCACAUUUGGCCUGGGCUGCUGGCAGGUAAAG (((((((((((((...(--((-(.......))))----------..........((((........))))....))))))...)))))))............ ( -14.30) >consensus AC__UCCUCUACCUACACAAA_CAACUAAUCAUUGUUGCC____ACCCAUUCCCCAGCUCAUACCGGCCACCACAUUUGGCUUGGGCUGCUGGCAAGUGAAG ...............................................((((..(((((.....(((((((.......)))).)))...)))))..))))... (-14.13 = -14.05 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:38 2006